| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,568,431 – 6,568,601 |
| Length | 170 |
| Max. P | 0.995564 |

| Location | 6,568,431 – 6,568,546 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.33 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -25.01 |
| Energy contribution | -24.97 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6568431 115 - 22224390 CAUGCAGCCAUGUAUCAUCAUCGAUCGCUGA-----UCGAUCUGUGGAUCGAGUGACACACGGGCCCGGACCCGGGGGCAAGGUGUUAAUUCAAUUACUUCUAUUAUGAAUAACAACAAA ..(((..((.(((.((((..((((((((.((-----....)).)).)))))))))).))).)).((((....)))).))).(.(((((.((((.............))))))))).)... ( -30.42) >DroSec_CAF1 13352 114 - 1 CAUGCAGCCAUGGAUCAUCAUCGAUCGCUCA-----UCGAUCUGUGGAUCGAGUGACACACGGGCCUGGACCCGG-GGCAAGGUGUUAAUUCAAUUACUUCUAUUAUGAAUAACAACAAA ((((....))))((((..(((.(((((....-----.))))).)))))))(((((((((...(.((((....)))-).)...))))).))))......(((......))).......... ( -30.00) >DroSim_CAF1 23908 114 - 1 CAUGCAGCCAUGGAUCAUCAUCGAUCGCUCA-----UCGAUCUGUGGAUCGAGUGACACACGGGCCUGGACCCGG-GGCAAGGUGUUAAUUCAAUUACUUCUAUUAUGAAUAACAACAAA ((((....))))((((..(((.(((((....-----.))))).)))))))(((((((((...(.((((....)))-).)...))))).))))......(((......))).......... ( -30.00) >DroEre_CAF1 13057 111 - 1 CAUGCAGCCAUGGAUCAUUAU---UCAGUGA-----UCGAUCUGUGGAUCGAGUGACACACGGGCCCGGACCCGG-GGCAAGGUGUUAAUUCAAUUACUUCUAUUAUGAAUAACAACAAA ((((...((((((((((((((---...))))-----).)))))))))...(((((((((...(.((((....)))-).)...))))).))))............))))............ ( -33.60) >DroYak_CAF1 22938 116 - 1 CAUGCAGCCAUGGAUCAUCGU---UCGAUGAAUCUGUGGAUCUGUGGAUUGAGUGACACACGGGCCCGGACCCGG-GGCAAAGUGUUAAUUCAAUUACUUCUAUUAUGAAUAACAACAAA (((((((((((((((((((..---..)))).))))))))..)))).(((((((((((((...(.((((....)))-).)...))))).)))))))).........)))............ ( -40.00) >consensus CAUGCAGCCAUGGAUCAUCAUCGAUCGCUGA_____UCGAUCUGUGGAUCGAGUGACACACGGGCCCGGACCCGG_GGCAAGGUGUUAAUUCAAUUACUUCUAUUAUGAAUAACAACAAA ((((...(((((((((......................)))))))))...(((((((((...(.((((....))).).)...))))).))))............))))............ (-25.01 = -24.97 + -0.04)

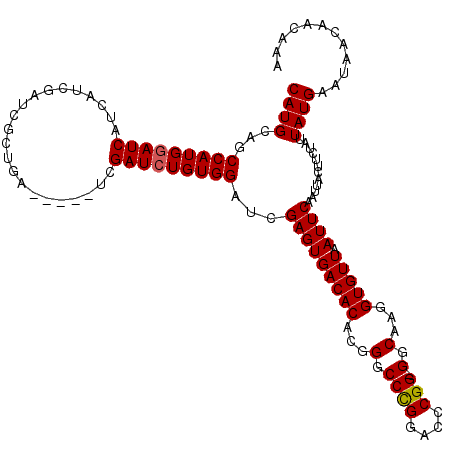

| Location | 6,568,471 – 6,568,574 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.84 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -27.46 |
| Energy contribution | -28.98 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.642011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6568471 103 + 22224390 UGCCCCCGGGUCCGGGCCCGUGUGUCACUCGAUCCACAGAUCGA-----UCAGCGAUCGAUGAUGAUACAUGGCUGCAUGGAGCGGUGGUAUCACCACAUCGAUUGCC------------ .((.(.(((((....))))).).))...((((((....))))))-----...((((((((((.((((((...(((((.....))))).))))))...)))))))))).------------ ( -42.50) >DroSec_CAF1 13392 102 + 1 UGCC-CCGGGUCCAGGCCCGUGUGUCACUCGAUCCACAGAUCGA-----UGAGCGAUCGAUGAUGAUCCAUGGCUGCAUGGAGCGGUGGUAUCACCACAUCGAUUGCC------------ ....-.(((((....)))))....(((.((((((....))))))-----)))((((((((((....((((((....))))))..((((....)))).)))))))))).------------ ( -45.10) >DroSim_CAF1 23948 102 + 1 UGCC-CCGGGUCCAGGCCCGUGUGUCACUCGAUCCACAGAUCGA-----UGAGCGAUCGAUGAUGAUCCAUGGCUGCAUGGAGCGGUAAUAUCACCACAUCGAUUGCC------------ ....-.(((((....)))))....(((.((((((....))))))-----)))((((((((((....((((((....))))))..(((......))).)))))))))).------------ ( -42.30) >DroEre_CAF1 13097 111 + 1 UGCC-CCGGGUCCGGGCCCGUGUGUCACUCGAUCCACAGAUCGA-----UCACUGA---AUAAUGAUCCAUGGCUGCAUGGAGCAGCAGUAUCACCUCAUCGAUUGACUAUUUUUGAAUG ....-.(((((....)))))...((((.((((((....))))((-----(.((((.---....((.((((((....)))))).)).)))))))........)).))))............ ( -32.50) >DroYak_CAF1 22978 116 + 1 UGCC-CCGGGUCCGGGCCCGUGUGUCACUCAAUCCACAGAUCCACAGAUUCAUCGA---ACGAUGAUCCAUGGCUGCAUGGAGCGCUAGUAUCACCUCAUCGAUUGGCUAUUAUUGAAUG .(((-.(((((....))))).(((..(((...(((((((..(((..(((.((((..---..)))))))..))))))..)))).....)))..)))..........)))............ ( -31.00) >consensus UGCC_CCGGGUCCGGGCCCGUGUGUCACUCGAUCCACAGAUCGA_____UCAGCGAUCGAUGAUGAUCCAUGGCUGCAUGGAGCGGUAGUAUCACCACAUCGAUUGCC____________ ......(((((....)))))........((((((....))))))........((((((((((.((((..((.(((((.....))))).))))))...))))))))))............. (-27.46 = -28.98 + 1.52)



| Location | 6,568,511 – 6,568,601 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -29.85 |
| Consensus MFE | -20.09 |
| Energy contribution | -19.90 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6568511 90 + 22224390 UCGAUCAGCGAUCGAUGAUGAUACAUGGCUGCAUGGAGCGGUGGUAUCACCACAUCGAUUGCC-------------------------UAUUGUUAUGGCUUAAUUUUAGUUGAA ....(((((((((((((.((((((...(((((.....))))).))))))...))))))))(((-------------------------.........))).........))))). ( -31.00) >DroSec_CAF1 13431 90 + 1 UCGAUGAGCGAUCGAUGAUGAUCCAUGGCUGCAUGGAGCGGUGGUAUCACCACAUCGAUUGCC-------------------------CAUUGUUGUGGCUUAAUUUUAGUUGGA .(((((.((((((((((....((((((....))))))..((((....)))).)))))))))).-------------------------)))))....((((.......))))... ( -33.90) >DroSim_CAF1 23987 90 + 1 UCGAUGAGCGAUCGAUGAUGAUCCAUGGCUGCAUGGAGCGGUAAUAUCACCACAUCGAUUGCC-------------------------CAUUGUUGUGGCUUAAUUUUAGUUGGA .(((((.((((((((((....((((((....))))))..(((......))).)))))))))).-------------------------)))))....((((.......))))... ( -31.10) >DroEre_CAF1 13136 112 + 1 UCGAUCACUGA---AUAAUGAUCCAUGGCUGCAUGGAGCAGCAGUAUCACCUCAUCGAUUGACUAUUUUUGAAUGGCUCUCGUUUGCUUAUUGUUAUGACUUCAAUUUAGUUGGA ..(((.((((.---....((.((((((....)))))).)).)))))))..........(..((((...(((((.(....((((..((.....)).))))))))))..))))..). ( -23.40) >consensus UCGAUCAGCGAUCGAUGAUGAUCCAUGGCUGCAUGGAGCGGUAGUAUCACCACAUCGAUUGCC_________________________CAUUGUUAUGGCUUAAUUUUAGUUGGA (((((..((((((((((.((((..((.(((((.....))))).))))))...))))))))))..........................(((....)))...........))))). (-20.09 = -19.90 + -0.19)

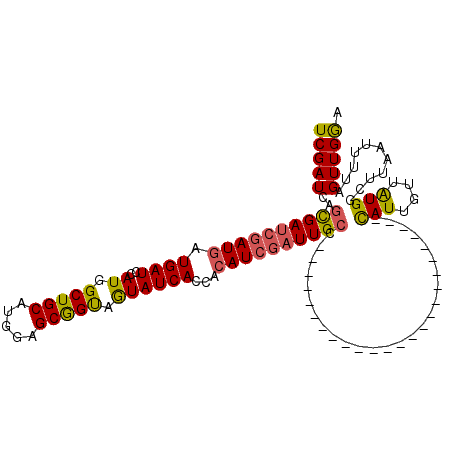

| Location | 6,568,511 – 6,568,601 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -16.41 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6568511 90 - 22224390 UUCAACUAAAAUUAAGCCAUAACAAUA-------------------------GGCAAUCGAUGUGGUGAUACCACCGCUCCAUGCAGCCAUGUAUCAUCAUCGAUCGCUGAUCGA ...........................-------------------------(((.(((((((..(((((((....(((......)))...)))))))))))))).)))...... ( -21.10) >DroSec_CAF1 13431 90 - 1 UCCAACUAAAAUUAAGCCACAACAAUG-------------------------GGCAAUCGAUGUGGUGAUACCACCGCUCCAUGCAGCCAUGGAUCAUCAUCGAUCGCUCAUCGA ......................(.(((-------------------------(((.((((((((((((....))))))((((((....)))))).....)))))).)))))).). ( -29.80) >DroSim_CAF1 23987 90 - 1 UCCAACUAAAAUUAAGCCACAACAAUG-------------------------GGCAAUCGAUGUGGUGAUAUUACCGCUCCAUGCAGCCAUGGAUCAUCAUCGAUCGCUCAUCGA ......................(.(((-------------------------(((.((((((((((((....))))))((((((....)))))).....)))))).)))))).). ( -27.50) >DroEre_CAF1 13136 112 - 1 UCCAACUAAAUUGAAGUCAUAACAAUAAGCAAACGAGAGCCAUUCAAAAAUAGUCAAUCGAUGAGGUGAUACUGCUGCUCCAUGCAGCCAUGGAUCAUUAU---UCAGUGAUCGA ....((((..(((((((....)).....((........))..)))))...))))(.((((.(((((((((...(((((.....))))).....)))))..)---))).)))).). ( -19.20) >consensus UCCAACUAAAAUUAAGCCACAACAAUA_________________________GGCAAUCGAUGUGGUGAUACCACCGCUCCAUGCAGCCAUGGAUCAUCAUCGAUCGCUCAUCGA ....................................................(((.(((((((.((((....))))(.((((((....)))))).)..))))))).)))...... (-16.41 = -16.48 + 0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:35:00 2006