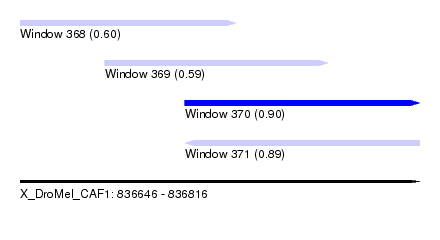
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 836,646 – 836,816 |
| Length | 170 |
| Max. P | 0.904383 |

| Location | 836,646 – 836,738 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 88.77 |
| Mean single sequence MFE | -19.15 |
| Consensus MFE | -14.94 |
| Energy contribution | -16.25 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 836646 92 + 22224390 UUCGCUACGCACAUUCAUGAAAAUCUCAUCGCAUAUAAUCAGUGCCACAUUCAGCGCAAAUCUCCAGGUACCACGCCCACUUCGCGGGCUUA ..((((..........((((.....)))).((((.......)))).......))))..................((((.......))))... ( -14.40) >DroSec_CAF1 37523 92 + 1 CGCCCUACGCACAUUCAUGAAAAUCUCAUGGCAUAUAAUCAGUGCCACAUUCAGCGCAAAUCUCCAGGUGCCACGCCCACCCCGCGGGCUUA .((((...((((..((((((.....))))))..........((((........))))..........))))...((.......))))))... ( -23.60) >DroSim_CAF1 33938 92 + 1 CGCCCUACGCACAUUCAUGAAAAUCUCAUGGCAUAUAAUCAGUGCCACAUUCAGCGCAAAUCUCCAGGUGCCACGCCCACCCCGCGGGCUUA .((((...((((..((((((.....))))))..........((((........))))..........))))...((.......))))))... ( -23.60) >DroYak_CAF1 34348 78 + 1 CUCCCUACGCACAUUCAUGAAAAUCUCAUGGCAUAUAAUCAGUGCCACAUUCAGCGCAAAUCUCCAGGUGCC--------------GGCUUA ...((...((((..((((((.....))))))..........((((........))))..........)))).--------------)).... ( -15.00) >consensus CGCCCUACGCACAUUCAUGAAAAUCUCAUGGCAUAUAAUCAGUGCCACAUUCAGCGCAAAUCUCCAGGUGCCACGCCCACCCCGCGGGCUUA ........((((..((((((.....))))))..........((((........))))..........))))...((((.......))))... (-14.94 = -16.25 + 1.31)

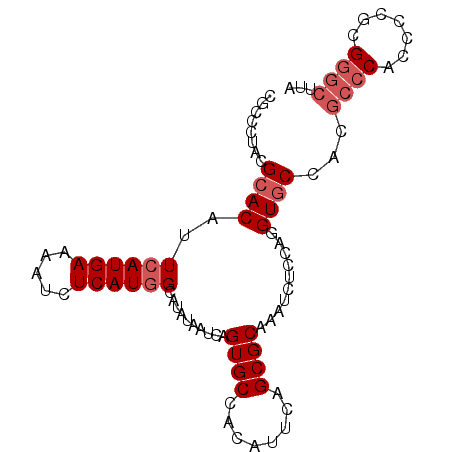
| Location | 836,682 – 836,777 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 89.35 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -18.09 |
| Energy contribution | -18.52 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 836682 95 + 22224390 AAUCAGUGCCACAUUCAGCGCAAAUCUCCAGGUACCACGCCCACUUCGCGGGCUUAUUC-CCACCCGCUAAGGCGGCAAACGCAUUACAUAUGCCA .....((((........)))).................(((..(((.(((((.......-...))))).)))..)))....((((.....)))).. ( -24.40) >DroSec_CAF1 37559 95 + 1 AAUCAGUGCCACAUUCAGCGCAAAUCUCCAGGUGCCACGCCCACCCCGCGGGCUUAUUC-CCACCCGCUAAGGCGGCAAACGCAUUACAUAUGCCA ......((((.......((((..........))))...(((......(((((.......-...)))))...)))))))...((((.....)))).. ( -24.60) >DroSim_CAF1 33974 95 + 1 AAUCAGUGCCACAUUCAGCGCAAAUCUCCAGGUGCCACGCCCACCCCGCGGGCUUACUC-CCACCCGCUAAGGCGGCAAACGCAUUACAUAUGCCA ......((((.......((((..........))))...(((......(((((.......-...)))))...)))))))...((((.....)))).. ( -24.60) >DroYak_CAF1 34384 82 + 1 AAUCAGUGCCACAUUCAGCGCAAAUCUCCAGGUGCC--------------GGCUUAUCCCCCGCCCGCUAAGGCGGCAAACGCAUUACAUAUGCCA .....((((........))))...........((((--------------(.((((.(........).)))).)))))...((((.....)))).. ( -19.80) >consensus AAUCAGUGCCACAUUCAGCGCAAAUCUCCAGGUGCCACGCCCACCCCGCGGGCUUAUUC_CCACCCGCUAAGGCGGCAAACGCAUUACAUAUGCCA .....((((........)))).........((((((.(((.......))))))...........((((....))))................))). (-18.09 = -18.52 + 0.44)

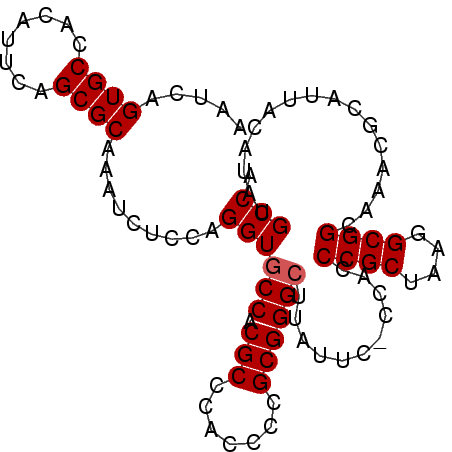
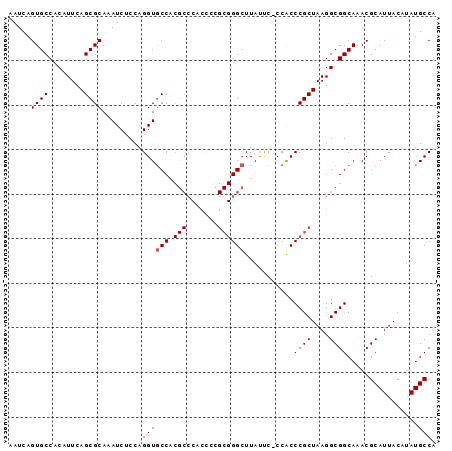
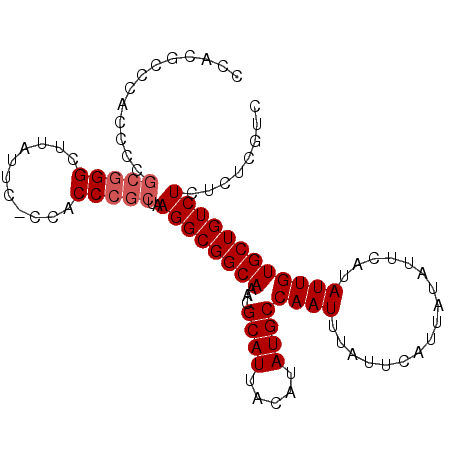
| Location | 836,716 – 836,816 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 89.88 |
| Mean single sequence MFE | -20.45 |
| Consensus MFE | -17.73 |
| Energy contribution | -18.48 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 836716 100 + 22224390 CCACGCCCACUUCGCGGGCUUAUUC-CCACCCGCUAAGGCGGCAAACGCAUUACAUAUGCCAAUUUAUUCAUUAUAUUCAUAUUGUGCUGUCUCUCUCGUC .((((((..(((.(((((.......-...))))).)))..)))....((((.....))))........................))).............. ( -21.90) >DroSec_CAF1 37593 100 + 1 CCACGCCCACCCCGCGGGCUUAUUC-CCACCCGCUAAGGCGGCAAACGCAUUACAUAUGCCAAUUUAUUCAUUAUAUUCAUAUUGUGCUGUCUCUCUCGUC ..(((........(((((.......-...)))))..((((((((...((((.....))))((((.................))))))))))))....))). ( -21.43) >DroSim_CAF1 34008 100 + 1 CCACGCCCACCCCGCGGGCUUACUC-CCACCCGCUAAGGCGGCAAACGCAUUACAUAUGCCAAUUUAUUCAUUAUAUUCAUAUUGUGCUGUCUCUCUCGUC ..(((........(((((.......-...)))))..((((((((...((((.....))))((((.................))))))))))))....))). ( -21.43) >DroYak_CAF1 34418 87 + 1 CC--------------GGCUUAUCCCCCGCCCGCUAAGGCGGCAAACGCAUUACAUAUGCCAAUUUAUUCAUUAUAUUCAUAUUGUGCUGUCUCGCUCGUC ..--------------(((.........))).((..((((((((...((((.....))))((((.................)))))))))))).))..... ( -17.03) >consensus CCACGCCCACCCCGCGGGCUUAUUC_CCACCCGCUAAGGCGGCAAACGCAUUACAUAUGCCAAUUUAUUCAUUAUAUUCAUAUUGUGCUGUCUCUCUCGUC .............(((((...........)))))..((((((((...((((.....))))((((.................))))))))))))........ (-17.73 = -18.48 + 0.75)

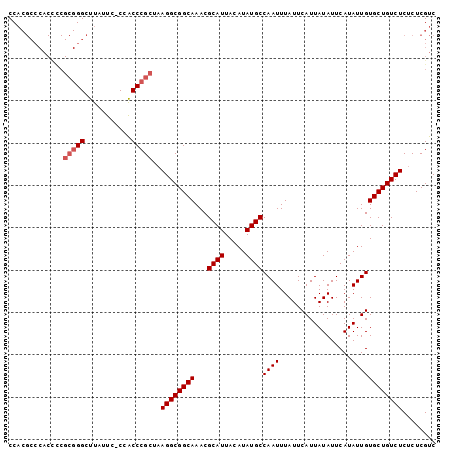
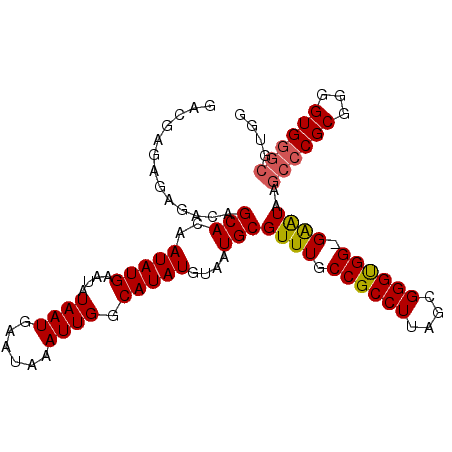
| Location | 836,716 – 836,816 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 89.88 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -22.67 |
| Energy contribution | -23.05 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 836716 100 - 22224390 GACGAGAGAGACAGCACAAUAUGAAUAUAAUGAAUAAAUUGGCAUAUGUAAUGCGUUUGCCGCCUUAGCGGGUGG-GAAUAAGCCCGCGAAGUGGGCGUGG ..(....)......(((.......................((((.(((.....))).))))((((..((((((..-......)))))).....))))))). ( -30.10) >DroSec_CAF1 37593 100 - 1 GACGAGAGAGACAGCACAAUAUGAAUAUAAUGAAUAAAUUGGCAUAUGUAAUGCGUUUGCCGCCUUAGCGGGUGG-GAAUAAGCCCGCGGGGUGGGCGUGG .(((....((((.(((..(((((....((((......)))).)))))....))))))).(((((((.((((((..-......))))))))))))).))).. ( -32.50) >DroSim_CAF1 34008 100 - 1 GACGAGAGAGACAGCACAAUAUGAAUAUAAUGAAUAAAUUGGCAUAUGUAAUGCGUUUGCCGCCUUAGCGGGUGG-GAGUAAGCCCGCGGGGUGGGCGUGG .(((....((((.(((..(((((....((((......)))).)))))....))))))).(((((((.((((((..-......))))))))))))).))).. ( -32.50) >DroYak_CAF1 34418 87 - 1 GACGAGCGAGACAGCACAAUAUGAAUAUAAUGAAUAAAUUGGCAUAUGUAAUGCGUUUGCCGCCUUAGCGGGCGGGGGAUAAGCC--------------GG ..((.((......(((..(((((....((((......)))).)))))....)))((((.((((((....)))))).))))..)))--------------). ( -25.00) >consensus GACGAGAGAGACAGCACAAUAUGAAUAUAAUGAAUAAAUUGGCAUAUGUAAUGCGUUUGCCGCCUUAGCGGGUGG_GAAUAAGCCCGCGGGGUGGGCGUGG .............(((..(((((....((((......)))).)))))....)))((((.((((((....)))))).))))..((((((...)))))).... (-22.67 = -23.05 + 0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:51 2006