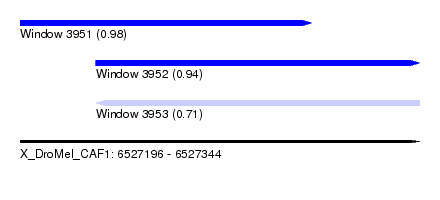
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,527,196 – 6,527,344 |
| Length | 148 |
| Max. P | 0.983349 |

| Location | 6,527,196 – 6,527,304 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.85 |
| Mean single sequence MFE | -32.21 |
| Consensus MFE | -20.12 |
| Energy contribution | -19.57 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
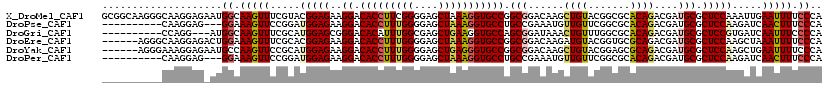
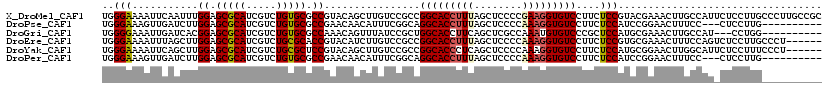
>X_DroMel_CAF1 6527196 108 + 22224390 UCUUUGC------------AGUGCGACAGUGGCGGUAAGGGCGGCAAGGGCAAGGAGAAUGGCAAGUUUCGUACGGAGAAGGACACCUUCGGGGAGCUAAAGGUGCCGGCGGACAAGCUG (((((((------------...((....)).((.((....)).))....)))))))....(((..(((.(((........((.((((((..(....)..)))))))).))))))..))). ( -31.10) >DroPse_CAF1 527 88 + 1 U-CAGGC------------A----AAAAAGGGCAAUUC------------CAAGGAG---GGAAAGUUCCGGAUGGAGAAGGACACCUUUGGGGAGCUAAAGGUGCCUGCCGAAAUGUUG .-..(((------------.----...........(((------------((.((((---......))))...))))).(((.(((((((((....)))))))))))))))......... ( -31.00) >DroSim_CAF1 476 96 + 1 UCUUUGC------------AGUGUGACAAGGGCGGUAAGGGCGGCAAGGGC------------AAGUUCCGCACGGAGAAGGACACCUUCGGGGAGCUAAAGGUGCCGGCGGACAAGCUG .((((((------------.....).)))))((.((....)).))...(((------------..((.((((.(((.......((((((..(....)..)))))))))))))))..))). ( -31.21) >DroEre_CAF1 529 90 + 1 UAUUUGC---------------------AGGGCGACA---------AGGGCAAGGAGACUGGAAAGUUUCGCACGGAGAAGGACACCUUUGGGGAGCUAAAGGUGCCGGCGGACAAGAUG ..(((((---------------------..(....).---------...)))))((((((....))))))((.(((.......(((((((((....)))))))))))))).......... ( -30.61) >DroYak_CAF1 541 111 + 1 UAUUUGCAUUUGCAUUCGCAGUGCGACAAGGGCGGCA---------AGGGAAAGGAGAAUGCCAAGUUCCGCAUGGAGAAGGACACCUUUGGGGAGCUGAGGGUGCCGGCGGACAAGCUG ...(((((((.((....)))))))))....(((((((---------.............))))..((.((((........((.((((((..(....)..)))))))).))))))..))). ( -38.32) >DroPer_CAF1 524 88 + 1 U-CAGGC------------A----AAAAAGGGCAAUUC------------CAAGGAG---GGAAAGUUCCGGAUGGAGAAGGACACCUUUGGGGAGCUAAAGGUGCCUGCCGAAAUGUUG .-..(((------------.----...........(((------------((.((((---......))))...))))).(((.(((((((((....)))))))))))))))......... ( -31.00) >consensus U_UUUGC____________A____GACAAGGGCGAUA_________AGGGCAAGGAG___GGAAAGUUCCGCACGGAGAAGGACACCUUUGGGGAGCUAAAGGUGCCGGCGGACAAGCUG ....(((...........................................................((((....))))..((.(((((((((....))))))))))).)))......... (-20.12 = -19.57 + -0.55)

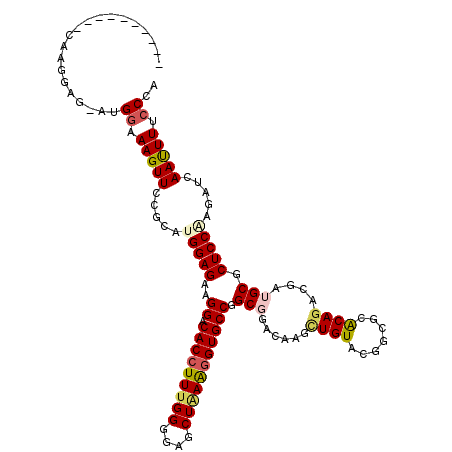
| Location | 6,527,224 – 6,527,344 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.91 |
| Mean single sequence MFE | -45.30 |
| Consensus MFE | -26.33 |
| Energy contribution | -26.67 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6527224 120 + 22224390 GCGGCAAGGGCAAGGAGAAUGGCAAGUUUCGUACGGAGAAGGACACCUUCGGGGAGCUAAAGGUGCCGGCGGACAAGCUGUACGGCGCACAGACGAUGCGCUCCAAAUUGAAUUUUCCCA ...((....))..(((((((..(((....(((((......((.((((((..(....)..))))))))(((......))))))))((((((....).)))))......))).))))))).. ( -39.30) >DroPse_CAF1 548 107 + 1 ----------CAAGGAG---GGAAAGUUCCGGAUGGAGAAGGACACCUUUGGGGAGCUAAAGGUGCCUGCCGAAAUGUUGUUCGGCGCACAGACGAUGCGCUCCAAGAUCAACUUUCCCA ----------......(---((((((((.....(((((.(((.(((((((((....))))))))))))((((((......))))))((((....).))).))))).....))))))))). ( -48.40) >DroGri_CAF1 933 107 + 1 ----------CCAGG---AUGGCAAGUUUCGCAUGGAGCGGGACACAUUUGGCGAGCUGAAGGUGCCAGCGGAUAAACUGUUUGGCGCACAGACGAUGCGCUCCGUGAUCAAUUUCCCCA ----------...((---..((.(((((...(((((((((.(.(...((..(....)..)).((((((((((.....))).)))))))......).).)))))))))...))))))))). ( -36.80) >DroEre_CAF1 545 114 + 1 ------AGGGCAAGGAGACUGGAAAGUUUCGCACGGAGAAGGACACCUUUGGGGAGCUAAAGGUGCCGGCGGACAAGAUGUACGGUGCGCAGACGAUGCGCUCCAAGCUAAAUUUUCCCA ------...((...((((((....))))))))..((((((((.(((((((((....)))))))))))((((.((.....)).)((.(((((.....))))).))..)))...)))))).. ( -45.10) >DroYak_CAF1 578 114 + 1 ------AGGGAAAGGAGAAUGCCAAGUUCCGCAUGGAGAAGGACACCUUUGGGGAGCUGAGGGUGCCGGCGGACAAGCUGUACGGAGCGCAGACGAUGCGCUCCAAGCUGAAUUUUCCCA ------.(((((((..(..((((...((((....))))..((.((((((..(....)..))))))))))))..).((((....((((((((.....)))))))).))))...))))))). ( -53.80) >DroPer_CAF1 545 107 + 1 ----------CAAGGAG---GGAAAGUUCCGGAUGGAGAAGGACACCUUUGGGGAGCUAAAGGUGCCUGCCGAAAUGUUGUUCGGCGCACAGACGAUGCGCUCCAAGAUCAACUUUCCCA ----------......(---((((((((.....(((((.(((.(((((((((....))))))))))))((((((......))))))((((....).))).))))).....))))))))). ( -48.40) >consensus __________CAAGGAG_AUGGAAAGUUCCGCAUGGAGAAGGACACCUUUGGGGAGCUAAAGGUGCCGGCGGACAAGCUGUACGGCGCACAGACGAUGCGCUCCAAGAUCAAUUUUCCCA ....................((.(((((.....(((((..((.(((((((((....))))))))))).(((......((((.......))))....))).))))).....))))).)).. (-26.33 = -26.67 + 0.33)

| Location | 6,527,224 – 6,527,344 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.91 |
| Mean single sequence MFE | -36.91 |
| Consensus MFE | -22.03 |
| Energy contribution | -22.25 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
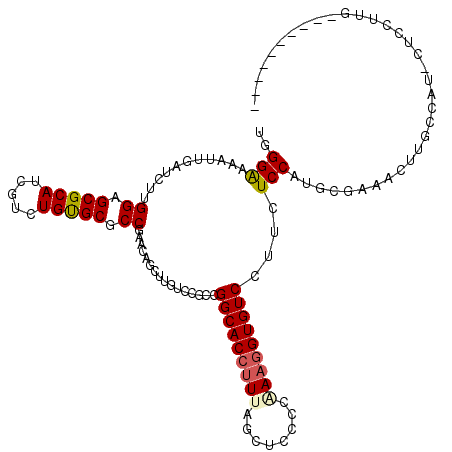
>X_DroMel_CAF1 6527224 120 - 22224390 UGGGAAAAUUCAAUUUGGAGCGCAUCGUCUGUGCGCCGUACAGCUUGUCCGCCGGCACCUUUAGCUCCCCGAAGGUGUCCUUCUCCGUACGAAACUUGCCAUUCUCCUUGCCCUUGCCGC .(((...........(((.((((((.....))))))(((((.((......)).(((((((((........))))))))).......))))).......))).........)))....... ( -33.85) >DroPse_CAF1 548 107 - 1 UGGGAAAGUUGAUCUUGGAGCGCAUCGUCUGUGCGCCGAACAACAUUUCGGCAGGCACCUUUAGCUCCCCAAAGGUGUCCUUCUCCAUCCGGAACUUUCC---CUCCUUG---------- .(((((((((.....(((((((((.....)))))((((((......)))))).(((((((((........)))))))))....)))).....))))))))---)......---------- ( -43.60) >DroGri_CAF1 933 107 - 1 UGGGGAAAUUGAUCACGGAGCGCAUCGUCUGUGCGCCAAACAGUUUAUCCGCUGGCACCUUCAGCUCGCCAAAUGUGUCCCGCUCCAUGCGAAACUUGCCAU---CCUGG---------- .((..((((((.....((.(((((.....))))).))...))))))..))(((((.....)))))...(((.(((.((..(((.....)))..))....)))---..)))---------- ( -26.20) >DroEre_CAF1 545 114 - 1 UGGGAAAAUUUAGCUUGGAGCGCAUCGUCUGCGCACCGUACAUCUUGUCCGCCGGCACCUUUAGCUCCCCAAAGGUGUCCUUCUCCGUGCGAAACUUUCCAGUCUCCUUGCCCU------ .(((.......((.((((((((((.....)))))..(((((.....(....).(((((((((........))))))))).......)))))......))))).)).....))).------ ( -33.30) >DroYak_CAF1 578 114 - 1 UGGGAAAAUUCAGCUUGGAGCGCAUCGUCUGCGCUCCGUACAGCUUGUCCGCCGGCACCCUCAGCUCCCCAAAGGUGUCCUUCUCCAUGCGGAACUUGGCAUUCUCCUUUCCCU------ .((((((....(((((((((((((.....)))))))))...)))).....(((((((((..............))))))....(((....)))....))).......)))))).------ ( -40.94) >DroPer_CAF1 545 107 - 1 UGGGAAAGUUGAUCUUGGAGCGCAUCGUCUGUGCGCCGAACAACAUUUCGGCAGGCACCUUUAGCUCCCCAAAGGUGUCCUUCUCCAUCCGGAACUUUCC---CUCCUUG---------- .(((((((((.....(((((((((.....)))))((((((......)))))).(((((((((........)))))))))....)))).....))))))))---)......---------- ( -43.60) >consensus UGGGAAAAUUGAUCUUGGAGCGCAUCGUCUGUGCGCCGAACAGCUUGUCCGCCGGCACCUUUAGCUCCCCAAAGGUGUCCUUCUCCAUGCGAAACUUGCCAU_CUCCUUG__________ ..(((...........((.(((((.....))))).))................(((((((((........)))))))))....))).................................. (-22.03 = -22.25 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:18 2006