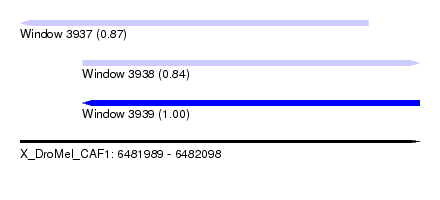
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,481,989 – 6,482,098 |
| Length | 109 |
| Max. P | 0.999135 |

| Location | 6,481,989 – 6,482,084 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 71.90 |
| Mean single sequence MFE | -13.64 |
| Consensus MFE | -3.93 |
| Energy contribution | -3.93 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6481989 95 - 22224390 GACCGAAAUCA-AAUGGCCAACACAAUAAAUUCACCUCUAAUGAAUUUUGAUUCAUUUCCAUUUGCUUUUGGCGCAAACAACUAAAUAAC----UCUAUG (.((((((.((-(((((..........(((((((.......)))))))..........))))))).))))))).................----...... ( -17.65) >DroVir_CAF1 53942 80 - 1 -----------AAAUGC-C-AAACAAUAAAUUG-GCACUAAUGAAU-UUGAUUCAUUUCGAUUUGCUUUUGGC---AAUAACUAAAUCAACAUUGC--GG -----------...(((-(-(((...(((((((-(....(((((((-...)))))))))))))))..))))))---)...................--.. ( -15.40) >DroPse_CAF1 43076 72 - 1 --------------------ACACAAUAAAUUCAGCACUAAUGAAUUUUGAUUCAUUUCCAUUCGCUUUC-GCAC-AACAGCCAAAUAAC----UU--CA --------------------..............((...(((((((....))))))).......((....-))..-....))........----..--.. ( -6.10) >DroEre_CAF1 35430 93 - 1 GACCAAAAUCA-AAUGGCCAACACAAUAAAUUCACCUCUAAUGAAUUUUGAUUCAUUUCCAUUUGCUUUUGGCGCAAACAACUAAAUAAC----UC--UG (.((((((.((-(((((..........(((((((.......)))))))..........))))))).))))))).................----..--.. ( -17.95) >DroAna_CAF1 54667 87 - 1 AGUUGAAAUCA-AAUGGCCAACACAAUAAAUUCAGCUCUAAUGAAUUUUGAUUCAUUUCCAUUUGCUUUUGGCGCAAACAACUAAAUA------------ .((..(((.((-(((((..........(((((((.......)))))))..........))))))).)))..))...............------------ ( -18.65) >DroPer_CAF1 45348 72 - 1 --------------------ACACAAUAAAUUCAGCACUAAUGAAUUUUGAUUCAUUUCCAUUCGCUUUC-GCAC-AACAGCCAAAUAAC----UU--CA --------------------..............((...(((((((....))))))).......((....-))..-....))........----..--.. ( -6.10) >consensus ____________AAUGG_C_ACACAAUAAAUUCAGCACUAAUGAAUUUUGAUUCAUUUCCAUUUGCUUUUGGCGC_AACAACUAAAUAAC____UC__CG .......................................(((((((....))))))).......((.....))........................... ( -3.93 = -3.93 + -0.00)

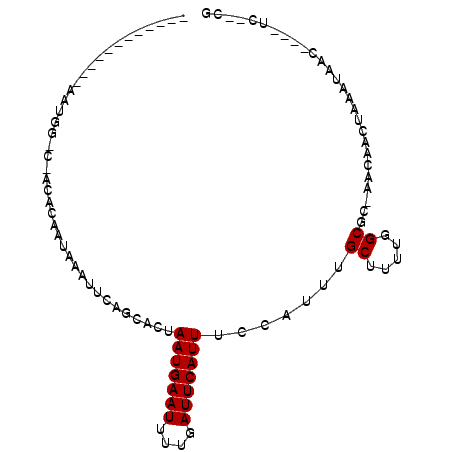

| Location | 6,482,006 – 6,482,098 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -19.65 |
| Energy contribution | -21.07 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6482006 92 + 22224390 GUUUGCGCCAAAAGCAAAUGGAAAUGAAUCAAAAUUCAUUAGAGGUGAAUUUAUUGUGUUGGCCAUUUGAUUUCGGUCCCAAGCGAGAUCAA ((((((.......))))))...(((((((....)))))))...(((.(((.......))).)))..(((((((((........))))))))) ( -22.80) >DroSec_CAF1 36221 92 + 1 GUUUGCGCCAAAAGCAAAUGGAAAUGAAUCAAAAUUCAUUAGAGGUGAAUUUAUUGUGUUGGCCAUUUGAUUUCGGUCCCAGUCGAGAUCAA ((((((.......))))))...(((((((....)))))))...(((.(((.......))).)))..((((((((((......)))))))))) ( -23.50) >DroSim_CAF1 34542 92 + 1 GUUUGCGCCAAAAGCAAAUGGAAAUGAAUCAAAAUUCAUUAGAGGUGAAUUUAUUGUGUUGGCCAUUUGAUUUCGGUCCCAGUCGAGAUCAA ((((((.......))))))...(((((((....)))))))...(((.(((.......))).)))..((((((((((......)))))))))) ( -23.50) >DroEre_CAF1 35445 92 + 1 GUUUGCGCCAAAAGCAAAUGGAAAUGAAUCAAAAUUCAUUAGAGGUGAAUUUAUUGUGUUGGCCAUUUGAUUUUGGUCCCAGCCGAGAUCAA ((((((.......))))))...(((((((....)))))))...(((.(((.......))).)))..(((((((((((....))))))))))) ( -25.30) >DroYak_CAF1 36072 92 + 1 GUUUGCGCCAAAAGCAAAUGGAAAUGAAUCAAAAUUCAUUAGAGGUGAAUUUAUUGUGUUGGCCAUUUGAUUUCGGUCCCAGCCGAGAUCAA ((((((.......))))))...(((((((....)))))))...(((.(((.......))).)))..(((((((((((....))))))))))) ( -27.40) >DroAna_CAF1 54676 85 + 1 GUUUGCGCCAAAAGCAAAUGGAAAUGAAUCAAAAUUCAUUAGAGCUGAAUUUAUUGUGUUGGCCAUUUGAUUUCAACUUCAACCG------- ((((((.......))))))((.(((((((....))))))).(((.((((.(((..(((.....))).))).)))).)))...)).------- ( -15.70) >consensus GUUUGCGCCAAAAGCAAAUGGAAAUGAAUCAAAAUUCAUUAGAGGUGAAUUUAUUGUGUUGGCCAUUUGAUUUCGGUCCCAGCCGAGAUCAA ...((.(((((..((((...((......)).((((((((.....)))))))).)))).)))))))..(((((((((......))))))))). (-19.65 = -21.07 + 1.42)

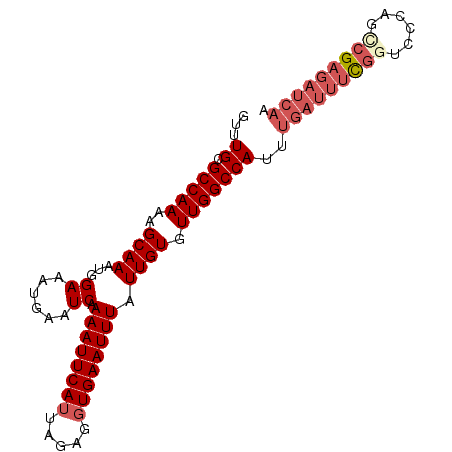
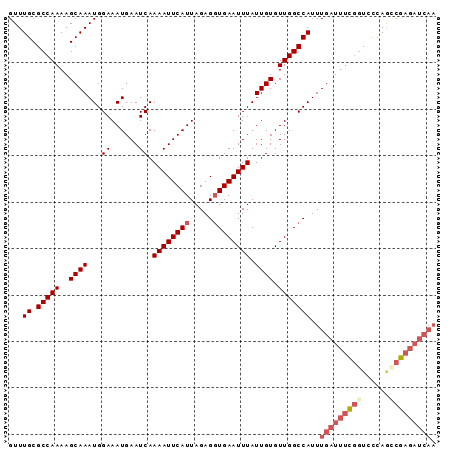
| Location | 6,482,006 – 6,482,098 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -20.22 |
| Consensus MFE | -18.86 |
| Energy contribution | -18.62 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6482006 92 - 22224390 UUGAUCUCGCUUGGGACCGAAAUCAAAUGGCCAACACAAUAAAUUCACCUCUAAUGAAUUUUGAUUCAUUUCCAUUUGCUUUUGGCGCAAAC ..........(((.(.((((((.(((((((..........(((((((.......)))))))..........))))))).))))))).))).. ( -21.05) >DroSec_CAF1 36221 92 - 1 UUGAUCUCGACUGGGACCGAAAUCAAAUGGCCAACACAAUAAAUUCACCUCUAAUGAAUUUUGAUUCAUUUCCAUUUGCUUUUGGCGCAAAC ...........((.(.((((((.(((((((..........(((((((.......)))))))..........))))))).))))))).))... ( -20.15) >DroSim_CAF1 34542 92 - 1 UUGAUCUCGACUGGGACCGAAAUCAAAUGGCCAACACAAUAAAUUCACCUCUAAUGAAUUUUGAUUCAUUUCCAUUUGCUUUUGGCGCAAAC ...........((.(.((((((.(((((((..........(((((((.......)))))))..........))))))).))))))).))... ( -20.15) >DroEre_CAF1 35445 92 - 1 UUGAUCUCGGCUGGGACCAAAAUCAAAUGGCCAACACAAUAAAUUCACCUCUAAUGAAUUUUGAUUCAUUUCCAUUUGCUUUUGGCGCAAAC ...........((.(.((((((.(((((((..........(((((((.......)))))))..........))))))).))))))).))... ( -20.45) >DroYak_CAF1 36072 92 - 1 UUGAUCUCGGCUGGGACCGAAAUCAAAUGGCCAACACAAUAAAUUCACCUCUAAUGAAUUUUGAUUCAUUUCCAUUUGCUUUUGGCGCAAAC ...........((.(.((((((.(((((((..........(((((((.......)))))))..........))))))).))))))).))... ( -20.15) >DroAna_CAF1 54676 85 - 1 -------CGGUUGAAGUUGAAAUCAAAUGGCCAACACAAUAAAUUCAGCUCUAAUGAAUUUUGAUUCAUUUCCAUUUGCUUUUGGCGCAAAC -------..(((...((..(((.(((((((..........(((((((.......)))))))..........))))))).)))..))...))) ( -19.35) >consensus UUGAUCUCGGCUGGGACCGAAAUCAAAUGGCCAACACAAUAAAUUCACCUCUAAUGAAUUUUGAUUCAUUUCCAUUUGCUUUUGGCGCAAAC ...........((.(.((((((.(((((((..........(((((((.......)))))))..........))))))).))))))).))... (-18.86 = -18.62 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:05 2006