
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,468,666 – 6,468,786 |
| Length | 120 |
| Max. P | 0.999025 |

| Location | 6,468,666 – 6,468,786 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.91 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -21.32 |
| Energy contribution | -21.28 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.999025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6468666 120 + 22224390 UUUCCAUUCAUACAUAUACAUGCAUAUAACCAAGUUAAGGUAGAUUGAAAAUAAGUAGUCCAGUGUUGUAUGCAAAUAAAUACUAUUACUUAUCAGCAGUACUUUUUGACACUUGGUUGU .........................(((((((((((((((...((((...(((((((((..((((((.(((....))))))))))))))))))...))))...)))))..)))))))))) ( -25.90) >DroSec_CAF1 22911 112 + 1 UUUCCAUUCAUACAUAU--------AUAACCAAGCUAAAGUAGAUUGAAAAUAAGUAGUCCAGUGUUGUAUGCAAAUAAUUACUACUACUUAUCAGCAAUACUUUUUGACACUUGGUUGU .................--------(((((((((.(((((...((((...(((((((((..((((...(((....)))..)))))))))))))...))))...)))))...))))))))) ( -25.50) >DroSim_CAF1 23372 112 + 1 UUUCCAUUCAUACAUAU--------AUAACCAAGCUAAAGUAGAUUGAAAAUAAGUAGUCCAGUGUUGUAUGCAAAUAAUUACUACUACUUAUCAGCACUACUUUUUGACACUUGGUUGU .................--------(((((((((..(((((((..((...(((((((((..((((...(((....)))..)))))))))))))...)))))))))......))))))))) ( -27.00) >DroEre_CAF1 22088 110 + 1 UCUUCGUUCAUA--UAU--------AUAACCAAGCUAAAGUAGAUUACUAAAAAGUAGUCCAGUGCUGUAUGCAAAUAAUUACUACUACUUAUCAGCACUACUCUUUGACACUUGGUUAU ............--...--------(((((((((.(((((..(((((((....))))))).(((((((.........................)))))))...)))))...))))))))) ( -25.61) >DroYak_CAF1 22649 108 + 1 UUUCCAUUCAUA----U--------GUAACCAAGUUAAAGUAGAGUAAAAAUAAGCAGUCCAGUGCUGUAUGCAAAUAAUUACUACUACUUAUCAGCACUACUUUUUGACAUUUGGUUAU ............----.--------((((((((((.(((((((.((....(((((.(((..((((...(((....)))..))))))).)))))..)).))))))).....)))))))))) ( -22.00) >consensus UUUCCAUUCAUACAUAU________AUAACCAAGCUAAAGUAGAUUGAAAAUAAGUAGUCCAGUGUUGUAUGCAAAUAAUUACUACUACUUAUCAGCACUACUUUUUGACACUUGGUUGU .........................(((((((((.(((((...((((...(((((((((..((((...(((....)))..)))))))))))))...))))...)))))...))))))))) (-21.32 = -21.28 + -0.04)


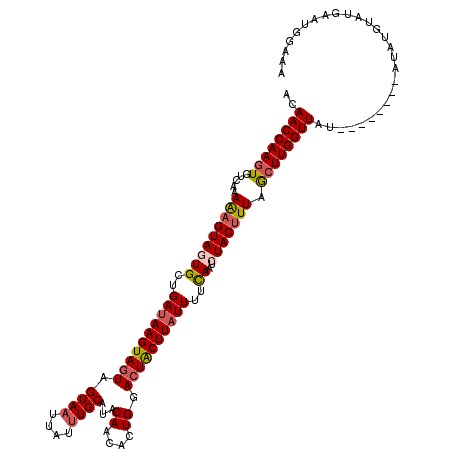
| Location | 6,468,666 – 6,468,786 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.91 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -23.08 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6468666 120 - 22224390 ACAACCAAGUGUCAAAAAGUACUGCUGAUAAGUAAUAGUAUUUAUUUGCAUACAACACUGGACUACUUAUUUUCAAUCUACCUUAACUUGGUUAUAUGCAUGUAUAUGUAUGAAUGGAAA ..((((((((........(((.(((.((((((((....)))))))).)))))).....((((.........))))..........)))))))).(((((((....)))))))........ ( -22.90) >DroSec_CAF1 22911 112 - 1 ACAACCAAGUGUCAAAAAGUAUUGCUGAUAAGUAGUAGUAAUUAUUUGCAUACAACACUGGACUACUUAUUUUCAAUCUACUUUAGCUUGGUUAU--------AUAUGUAUGAAUGGAAA ..((((((((.....(((((((((..((((((((((.((((....))))...((....)).))))))))))..)))..)))))).))))))))..--------................. ( -26.80) >DroSim_CAF1 23372 112 - 1 ACAACCAAGUGUCAAAAAGUAGUGCUGAUAAGUAGUAGUAAUUAUUUGCAUACAACACUGGACUACUUAUUUUCAAUCUACUUUAGCUUGGUUAU--------AUAUGUAUGAAUGGAAA ..((((((((.....(((((((((..((((((((((.((((....))))...((....)).))))))))))..))..))))))).))))))))..--------................. ( -28.50) >DroEre_CAF1 22088 110 - 1 AUAACCAAGUGUCAAAGAGUAGUGCUGAUAAGUAGUAGUAAUUAUUUGCAUACAGCACUGGACUACUUUUUAGUAAUCUACUUUAGCUUGGUUAU--------AUA--UAUGAACGAAGA ((((((((((.....(((((((((((((.(((((((.((((....))))...((....)).))))))).))))))..))))))).))))))))))--------...--............ ( -33.10) >DroYak_CAF1 22649 108 - 1 AUAACCAAAUGUCAAAAAGUAGUGCUGAUAAGUAGUAGUAAUUAUUUGCAUACAGCACUGGACUGCUUAUUUUUACUCUACUUUAACUUGGUUAC--------A----UAUGAAUGGAAA .(((((((..((...(((((((.(..((((((((((.((((....))))...((....)).))))))))))....).))))))).))))))))).--------.----............ ( -24.40) >consensus ACAACCAAGUGUCAAAAAGUAGUGCUGAUAAGUAGUAGUAAUUAUUUGCAUACAACACUGGACUACUUAUUUUCAAUCUACUUUAGCUUGGUUAU________AUAUGUAUGAAUGGAAA ..((((((((.....(((((((((..((((((((((.((((....))))...((....)).))))))))))..))..))))))).))))))))........................... (-23.08 = -23.68 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:34:00 2006