
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,468,084 – 6,468,176 |
| Length | 92 |
| Max. P | 0.557338 |

| Location | 6,468,084 – 6,468,176 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 84.45 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -17.35 |
| Energy contribution | -18.13 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
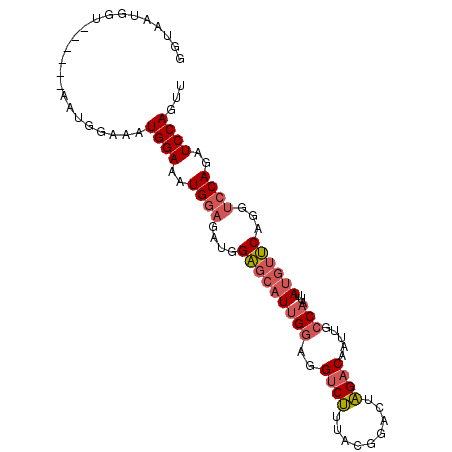
>X_DroMel_CAF1 6468084 92 + 22224390 GGUAAUGGU------AAUGGAAAUGGAAAUGGAGAUGGAGCAUUGGAGGUCUUUACGGACUAGACAAUUGCCAAUUAUGUUCAGGUCCAGAUCCAGUU .........------..((((..((((..((((.((((.((((((..(((((....)))))...))).)))...)))).))))..))))..))))... ( -26.10) >DroSec_CAF1 22326 92 + 1 GGUAAUGGU------AAUGGAAAUGGAAAUGGAGAUGGAGCAUUGGAGGUCUUUACGGACUAGACAAUUGCCAAUUAUGUUCAGGUCCAGAUCCAGUU .........------..((((..((((..((((.((((.((((((..(((((....)))))...))).)))...)))).))))..))))..))))... ( -26.10) >DroSim_CAF1 22801 98 + 1 GGUAAUGGUAAUGGAAAUGGAAAUGGAAAUGGAGAUGGAGCAUUGGAGGUCUUUGCGGACUAGACAAUUGCCAAUUAUGUUCAGGUCCAGAUCCAGUU .................((((..((((..((((.((((.((((((..(((((....)))))...))).)))...)))).))))..))))..))))... ( -26.10) >DroEre_CAF1 21536 98 + 1 GGUAAUGCUAAUGCCAAUGGAAAUGGAAAUGGAGAUGGAGCAUUGGAGGUCUUUACGGACUAGACAAUUGCCAAUUAUGUUCAGGUCCAGAUCCAGUU ((((.......))))..((((..((((..((((.((((.((((((..(((((....)))))...))).)))...)))).))))..))))..))))... ( -29.00) >DroYak_CAF1 22104 86 + 1 GGUAA------------UGGAAAUGGAAAUGGAGAUGGAGCAUUGGAGGUCUUUACGGACUAGACAAUUGCCAAUUAUGUUCAGGUCCAGAUCCAGUU .....------------((((..((((..((((.((((.((((((..(((((....)))))...))).)))...)))).))))..))))..))))... ( -26.10) >DroAna_CAF1 27755 83 + 1 AGGAAUGGU------AAUGCCAAUGGAAAUGGAGAUGGU--CUAGAAGGCCC-C-----GUGGACAAUUGCCAAUUAUGAGCAAA-GCAGAUCCAGUU .(((.((((------(((.(((.(((..........(((--(.....)))))-)-----))))...))))))).......((...-))...))).... ( -19.70) >consensus GGUAAUGGU______AAUGGAAAUGGAAAUGGAGAUGGAGCAUUGGAGGUCUUUACGGACUAGACAAUUGCCAAUUAUGUUCAGGUCCAGAUCCAGUU .......................((((..((((....(((((((((..((((.........)))).....)))...))))))...))))..))))... (-17.35 = -18.13 + 0.78)

| Location | 6,468,084 – 6,468,176 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 84.45 |
| Mean single sequence MFE | -15.58 |
| Consensus MFE | -13.29 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6468084 92 - 22224390 AACUGGAUCUGGACCUGAACAUAAUUGGCAAUUGUCUAGUCCGUAAAGACCUCCAAUGCUCCAUCUCCAUUUCCAUUUCCAUU------ACCAUUACC ...((((..((((..((......(((((.....((((.........))))..)))))....))..))))..))))........------......... ( -15.70) >DroSec_CAF1 22326 92 - 1 AACUGGAUCUGGACCUGAACAUAAUUGGCAAUUGUCUAGUCCGUAAAGACCUCCAAUGCUCCAUCUCCAUUUCCAUUUCCAUU------ACCAUUACC ...((((..((((..((......(((((.....((((.........))))..)))))....))..))))..))))........------......... ( -15.70) >DroSim_CAF1 22801 98 - 1 AACUGGAUCUGGACCUGAACAUAAUUGGCAAUUGUCUAGUCCGCAAAGACCUCCAAUGCUCCAUCUCCAUUUCCAUUUCCAUUUCCAUUACCAUUACC ...((((..((((..((......(((((.....((((.........))))..)))))....))..))))..))))....................... ( -15.70) >DroEre_CAF1 21536 98 - 1 AACUGGAUCUGGACCUGAACAUAAUUGGCAAUUGUCUAGUCCGUAAAGACCUCCAAUGCUCCAUCUCCAUUUCCAUUUCCAUUGGCAUUAGCAUUACC ....(((.((((((.....((....))......)))))))))............((((((......(((.............)))....))))))... ( -16.82) >DroYak_CAF1 22104 86 - 1 AACUGGAUCUGGACCUGAACAUAAUUGGCAAUUGUCUAGUCCGUAAAGACCUCCAAUGCUCCAUCUCCAUUUCCAUUUCCA------------UUACC ...((((..((((..((......(((((.....((((.........))))..)))))....))..))))..))))......------------..... ( -15.70) >DroAna_CAF1 27755 83 - 1 AACUGGAUCUGC-UUUGCUCAUAAUUGGCAAUUGUCCAC-----G-GGGCCUUCUAG--ACCAUCUCCAUUUCCAUUGGCAUU------ACCAUUCCU ...(((((....-.((((.((....))))))..))))).-----(-(.(((......--..................)))...------.))...... ( -13.86) >consensus AACUGGAUCUGGACCUGAACAUAAUUGGCAAUUGUCUAGUCCGUAAAGACCUCCAAUGCUCCAUCUCCAUUUCCAUUUCCAUU______ACCAUUACC ...((((..((((..((......(((((.....((((.........))))..)))))....))..))))..))))....................... (-13.29 = -13.40 + 0.11)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:58 2006