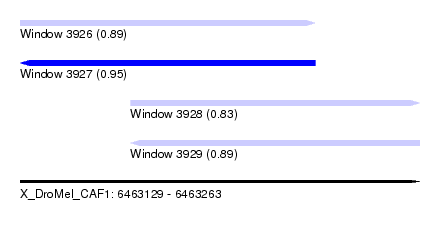
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,463,129 – 6,463,263 |
| Length | 134 |
| Max. P | 0.946112 |

| Location | 6,463,129 – 6,463,228 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -18.43 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6463129 99 + 22224390 CAGUUGCUCCAUCCGGUUGCAGUUGCAUGUUGCAUAC---------------------UGCAUAUGGCAGAUUGCACAUUGCACAUUACACGUUGCACGUUGCACAUAUGUUUGUAUGUU ((..(((.((....))..)))..))......((((((---------------------.(((((((((((..((((...((.......))...))))..)))).)))))))..)))))). ( -28.20) >DroSec_CAF1 17506 120 + 1 CAGUUGCUCCAUCCGGUUGCAGUUGCAUGUUGCAUACUGCAUAUUGCAUACUGCAUACUGCAUAUGGCAGAUUGCACAUUGCACAUUACACGCUGCACGUUGCACAUAUGUUUGUACGUU .............((..((((((((((...(((.....)))...)))).))))))(((.(((((((((((..((((....((.........))))))..)))).)))))))..))))).. ( -34.50) >DroSim_CAF1 17988 113 + 1 CAGUUGCUCCAUCCGGUUGCAGUUGCAUGUUGCAUACUGCA-------UACUGCAUACUGCAUAUGGCAGAUUGCACAUUGCACAUUACACGCUGCACGUUGCACAUAUGUUUGUAUGUU .............(((((((((((((.....))).))))))-------.))))(((((.(((((((((((..((((....((.........))))))..)))).)))))))..))))).. ( -35.30) >DroEre_CAF1 16758 95 + 1 CAGUUGCUCCAUCCGGUUGCAGUUGCAUGUUGCAUACU--------------GCAUACUGCAUAUGGCAGAUUGCACAUUGCACAU-------UGCACGUUGCACAUAUGU----GUGUU (((((...((((.(((((((((((((.....))).)))--------------))).))))...))))..)))))......((((((-------(((.....)))...))))----))... ( -28.40) >DroYak_CAF1 17546 81 + 1 CAGUUGCUCCAUCCGGUUGCAGUUGCAUGUUGCAUAC---------------------UGCAAAUGGCAGAUUGCAC-------AU-------UACACGUUGCACAUAUGU----GUGUU ..((.((.....(((.((((((((((.....))).))---------------------))))).)))......))))-------..-------(((((((.......))))----))).. ( -23.50) >consensus CAGUUGCUCCAUCCGGUUGCAGUUGCAUGUUGCAUACU______________GCAUACUGCAUAUGGCAGAUUGCACAUUGCACAUUACACG_UGCACGUUGCACAUAUGUUUGUAUGUU ...((((.((....))..)))).(((.....))).........................(((((((((((..((((.................))))..)))).)))))))......... (-18.43 = -18.83 + 0.40)



| Location | 6,463,129 – 6,463,228 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -18.49 |
| Energy contribution | -19.28 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6463129 99 - 22224390 AACAUACAAACAUAUGUGCAACGUGCAACGUGUAAUGUGCAAUGUGCAAUCUGCCAUAUGCA---------------------GUAUGCAACAUGCAACUGCAACCGGAUGGAGCAACUG ..........((((((.(((...((((...(((.....)))...))))...)))))))))((---------------------((.(((....(((....))).((....)).))))))) ( -26.40) >DroSec_CAF1 17506 120 - 1 AACGUACAAACAUAUGUGCAACGUGCAGCGUGUAAUGUGCAAUGUGCAAUCUGCCAUAUGCAGUAUGCAGUAUGCAAUAUGCAGUAUGCAACAUGCAACUGCAACCGGAUGGAGCAACUG ..(((((...((((((.(((...((((...(((.....)))...))))...)))))))))..)))))((((.(((....((((((.(((.....))))))))).((....)).))))))) ( -38.30) >DroSim_CAF1 17988 113 - 1 AACAUACAAACAUAUGUGCAACGUGCAGCGUGUAAUGUGCAAUGUGCAAUCUGCCAUAUGCAGUAUGCAGUA-------UGCAGUAUGCAACAUGCAACUGCAACCGGAUGGAGCAACUG ..(((((...((((((.(((...((((...(((.....)))...))))...)))))))))..)))))((((.-------((((((.(((.....))))))))).((....))....)))) ( -34.70) >DroEre_CAF1 16758 95 - 1 AACAC----ACAUAUGUGCAACGUGCA-------AUGUGCAAUGUGCAAUCUGCCAUAUGCAGUAUGC--------------AGUAUGCAACAUGCAACUGCAACCGGAUGGAGCAACUG ....(----((((.((..((.......-------.))..))))))).....(((..(((.(.((.(((--------------(((.(((.....))))))))))).).)))..))).... ( -27.20) >DroYak_CAF1 17546 81 - 1 AACAC----ACAUAUGUGCAACGUGUA-------AU-------GUGCAAUCUGCCAUUUGCA---------------------GUAUGCAACAUGCAACUGCAACCGGAUGGAGCAACUG ..(((----(.(((((.....))))).-------.)-------)))...((((((..(((((---------------------((.(((.....))))))))))..)).))))....... ( -21.30) >consensus AACAUACAAACAUAUGUGCAACGUGCA_CGUGUAAUGUGCAAUGUGCAAUCUGCCAUAUGCAGUAUGC______________AGUAUGCAACAUGCAACUGCAACCGGAUGGAGCAACUG ..........((((((.(((...((((...(((.....)))...))))...))))))))).......................((.(((....(((....))).((....)).))))).. (-18.49 = -19.28 + 0.79)

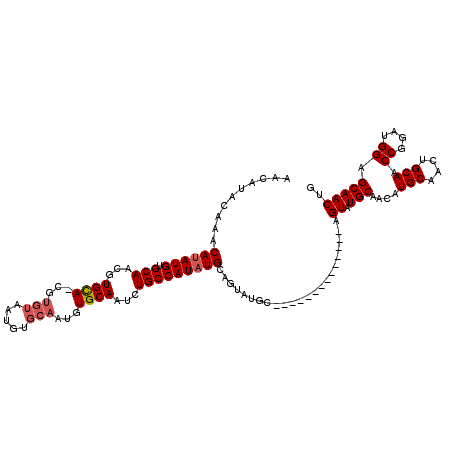
| Location | 6,463,166 – 6,463,263 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -25.46 |
| Consensus MFE | -16.73 |
| Energy contribution | -17.37 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6463166 97 + 22224390 ------------------UGCAUAUGGCAGAUUGCACAUUGCACAUUACACGUUGCACGUUGCACAUAUGUUUGUAUGUUGCAAGGCUCAAGUGUUUAUAUGUGACUGCUUUUUU ------------------.......(((((...((.....))((((.((((.(((..(.((((((((((....))))).))))).)..)))))))....))))..)))))..... ( -26.10) >DroSec_CAF1 17546 115 + 1 AUAUUGCAUACUGCAUACUGCAUAUGGCAGAUUGCACAUUGCACAUUACACGCUGCACGUUGCACAUAUGUUUGUACGUUGCAAGGCUCAAGUGUUUAUAUGUGACUGCUUUUCU .....(((...((((..((((.....))))..)))).....(((((.((((..((..(.(((((...((((....))))))))).)..)).))))....)))))..)))...... ( -28.60) >DroSim_CAF1 18028 108 + 1 A-------UACUGCAUACUGCAUAUGGCAGAUUGCACAUUGCACAUUACACGCUGCACGUUGCACAUAUGUUUGUAUGUUGCAAGGCUCAAGUGUUUAUAUGCGACUGCUUUUUU .-------...((((..((((.....))))..))))..(((((.((.((((..((..(.((((((((((....))))).))))).)..)).))))..)).))))).......... ( -29.40) >DroEre_CAF1 16796 92 + 1 ------------GCAUACUGCAUAUGGCAGAUUGCACAUUGCACAU-------UGCACGUUGCACAUAUGU----GUGUUGCAAGGCUCAAGUGUUUAUAUGUGACUGCUUUUUU ------------(((..((((.....))))....(((((.((((.(-------((..(.((((((((....----))).))))).)..)))))))....)))))..)))...... ( -26.20) >DroYak_CAF1 17583 77 + 1 ------------------UGCAAAUGGCAGAUUGCAC-------AU-------UACACGUUGCACAUAUGU----GUGUUGCAAGGCUCAAGUGUUUAUAUGUUUCU--UUUUUU ------------------.(((..((((...(((((.-------..-------(((((((.......))))----))).))))).)).))..)))............--...... ( -17.00) >consensus ____________GCAUACUGCAUAUGGCAGAUUGCACAUUGCACAUUACACG_UGCACGUUGCACAUAUGUUUGUAUGUUGCAAGGCUCAAGUGUUUAUAUGUGACUGCUUUUUU ...................(((((((((((..((((.................))))..)))).)))))))......((((((((((......))))...))))))......... (-16.73 = -17.37 + 0.64)

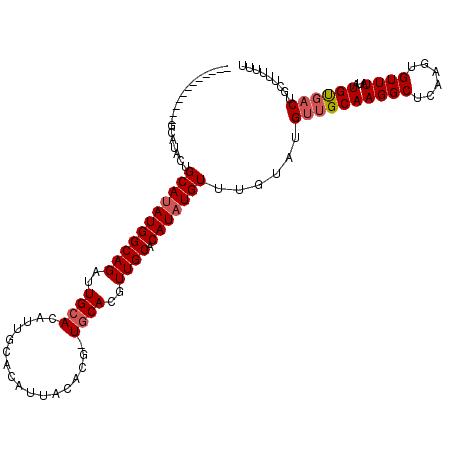

| Location | 6,463,166 – 6,463,263 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -12.37 |
| Energy contribution | -13.16 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6463166 97 - 22224390 AAAAAAGCAGUCACAUAUAAACACUUGAGCCUUGCAACAUACAAACAUAUGUGCAACGUGCAACGUGUAAUGUGCAAUGUGCAAUCUGCCAUAUGCA------------------ ......((((.(((((....(((((((.((.(((((.((((......))))))))).)).))).)))).)))))).(((.((.....))))).))).------------------ ( -24.70) >DroSec_CAF1 17546 115 - 1 AGAAAAGCAGUCACAUAUAAACACUUGAGCCUUGCAACGUACAAACAUAUGUGCAACGUGCAGCGUGUAAUGUGCAAUGUGCAAUCUGCCAUAUGCAGUAUGCAGUAUGCAAUAU ......((((.(((((....(((((((.((.(((((.((((......))))))))).)).))).)))).))))))....((((..((((.....))))..))))...)))..... ( -30.00) >DroSim_CAF1 18028 108 - 1 AAAAAAGCAGUCGCAUAUAAACACUUGAGCCUUGCAACAUACAAACAUAUGUGCAACGUGCAGCGUGUAAUGUGCAAUGUGCAAUCUGCCAUAUGCAGUAUGCAGUA-------U ......(((...((((((..(((((((.((.(((((.((((......))))))))).)).))).)))).))))))..)))(((..((((.....))))..)))....-------. ( -30.10) >DroEre_CAF1 16796 92 - 1 AAAAAAGCAGUCACAUAUAAACACUUGAGCCUUGCAACAC----ACAUAUGUGCAACGUGCA-------AUGUGCAAUGUGCAAUCUGCCAUAUGCAGUAUGC------------ ......((((.(((((....(((.(((.((.(((((.((.----.....))))))).)).))-------))))...))))))...((((.....))))..)))------------ ( -19.80) >DroYak_CAF1 17583 77 - 1 AAAAAA--AGAAACAUAUAAACACUUGAGCCUUGCAACAC----ACAUAUGUGCAACGUGUA-------AU-------GUGCAAUCUGCCAUUUGCA------------------ ......--............(((.(((.((.(((((.((.----.....))))))).)).))-------))-------))((((........)))).------------------ ( -11.00) >consensus AAAAAAGCAGUCACAUAUAAACACUUGAGCCUUGCAACAUACAAACAUAUGUGCAACGUGCA_CGUGUAAUGUGCAAUGUGCAAUCUGCCAUAUGCAGUAUGC____________ .............................................((((((.(((...((((...(((.....)))...))))...))))))))).................... (-12.37 = -13.16 + 0.79)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:57 2006