
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,456,276 – 6,456,372 |
| Length | 96 |
| Max. P | 0.709412 |

| Location | 6,456,276 – 6,456,372 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -16.86 |
| Energy contribution | -16.33 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6456276 96 + 22224390 CCGAAGUAACCAUCGCAUUUUUUUUUUUUUUGC----AAUCUUUAGCGAUCGCCAAGCAGGCGUGAAAUCAUCGUUAUUAUGAUGAUUAGU-UUUUUGCUC ....(((((...((((.............((((----........))))..(((.....)))))))(((((((((....)))))))))...-...))))). ( -21.70) >DroPse_CAF1 12463 89 + 1 UCAGAUUUGGCAUAGC---U---------UUGCGGCUGAUAUUUUCCUAUCGCAAAGCAGGCGUGAACUCAUUGCUAAUAUGAUGGUUAUUCUUUUUGCCC ........((((..((---(---------((((((..(........)..))))))))).((((((....)).))))....................)))). ( -22.50) >DroSec_CAF1 10760 87 + 1 CCGAAGUUACCAUCGCAUUU---------UUGC----GAUCUUUAGCGAUCGCCAAGCAGGCGUGAAAUCAUCGUUAUUAUGAUGAUUAGU-UUUUUGCUU ..((((.....((((((...---------.)))----)))))))(((((.((((.....))))...(((((((((....)))))))))...-...))))). ( -25.10) >DroSim_CAF1 10740 87 + 1 ACGAAGUUACCAUCGCAUUU---------UUGC----GAUCUUUAGCGAUCGCCAAGCAGGCGUGAAAUCAUCGUUAUUAUGAUGAUUAGU-UUUUUGCUC ..((((.....((((((...---------.)))----)))))))(((((.((((.....))))...(((((((((....)))))))))...-...))))). ( -25.10) >DroEre_CAF1 10020 87 + 1 GCAGAGUUACCAUCGCAUUU---------UCGC----GAUCUUUAGCGAUCGCCAAGCAGGCGUGAAAUCAUCGUUAUUAUGAUGAUUAGU-UUUUUGCUC ((((((..((..((((....---------((((----........))))..(((.....)))))))(((((((((....))))))))).))-.)))))).. ( -27.60) >DroYak_CAF1 10999 87 + 1 GCUGAGUUACCAUCACUUUU---------UUGC----GAUCUUUAGCGAUCGCCAAGCAGGCGUGAAAUCAUCGUUAUUAUGAUGAUUAGU-UUUUUGCUC ...((((.....((((....---------....----((((......))))(((.....)))))))(((((((((....)))))))))...-.....)))) ( -23.40) >consensus CCGAAGUUACCAUCGCAUUU_________UUGC____GAUCUUUAGCGAUCGCCAAGCAGGCGUGAAAUCAUCGUUAUUAUGAUGAUUAGU_UUUUUGCUC .((((...((..((((.....................((((......))))(((.....)))))))(((((((((....))))))))).))...))))... (-16.86 = -16.33 + -0.52)

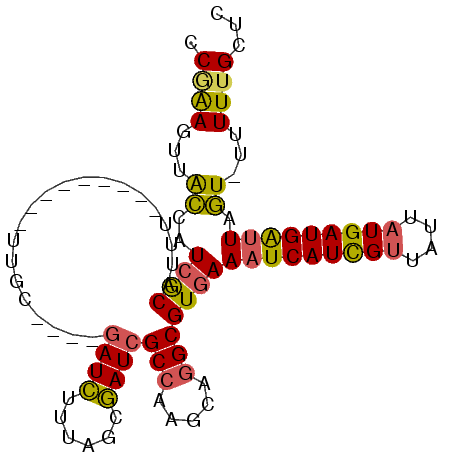
| Location | 6,456,276 – 6,456,372 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 83.49 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -14.64 |
| Energy contribution | -14.64 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6456276 96 - 22224390 GAGCAAAAA-ACUAAUCAUCAUAAUAACGAUGAUUUCACGCCUGCUUGGCGAUCGCUAAAGAUU----GCAAAAAAAAAAAAAAUGCGAUGGUUACUUCGG (((((....-...(((((((........))))))).......))))).((((((......))))----))................(((.(....).))). ( -21.54) >DroPse_CAF1 12463 89 - 1 GGGCAAAAAGAAUAACCAUCAUAUUAGCAAUGAGUUCACGCCUGCUUUGCGAUAGGAAAAUAUCAGCCGCAA---------A---GCUAUGCCAAAUCUGA .((((....((((.....((((.......))))))))......((((((((....((.....))...)))))---------)---))..))))........ ( -20.20) >DroSec_CAF1 10760 87 - 1 AAGCAAAAA-ACUAAUCAUCAUAAUAACGAUGAUUUCACGCCUGCUUGGCGAUCGCUAAAGAUC----GCAA---------AAAUGCGAUGGUAACUUCGG (((((....-...(((((((........))))))).......))))).((((((......))))----))..---------.....(((.(....).))). ( -23.24) >DroSim_CAF1 10740 87 - 1 GAGCAAAAA-ACUAAUCAUCAUAAUAACGAUGAUUUCACGCCUGCUUGGCGAUCGCUAAAGAUC----GCAA---------AAAUGCGAUGGUAACUUCGU (((((....-...(((((((........))))))).......))))).((((((......))))----))..---------....((((.(....).)))) ( -24.34) >DroEre_CAF1 10020 87 - 1 GAGCAAAAA-ACUAAUCAUCAUAAUAACGAUGAUUUCACGCCUGCUUGGCGAUCGCUAAAGAUC----GCGA---------AAAUGCGAUGGUAACUCUGC (((......-...(((((((........)))))))((((((.(..((.((((((......))))----)).)---------).).))).)))...)))... ( -23.00) >DroYak_CAF1 10999 87 - 1 GAGCAAAAA-ACUAAUCAUCAUAAUAACGAUGAUUUCACGCCUGCUUGGCGAUCGCUAAAGAUC----GCAA---------AAAAGUGAUGGUAACUCAGC (((......-...(((((((........)))))))....(((.((((.((((((......))))----))..---------..))))...)))..)))... ( -24.00) >consensus GAGCAAAAA_ACUAAUCAUCAUAAUAACGAUGAUUUCACGCCUGCUUGGCGAUCGCUAAAGAUC____GCAA_________AAAUGCGAUGGUAACUCCGC (((((........(((((((........))))))).......))))).((.(((((.............................))))).))........ (-14.64 = -14.64 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:51 2006