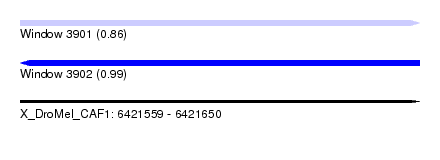
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,421,559 – 6,421,650 |
| Length | 91 |
| Max. P | 0.992668 |

| Location | 6,421,559 – 6,421,650 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.21 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.95 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6421559 91 + 22224390 -UUGGUCAACGUGUCGUAUGCGCACUUUGUAUAUUAUAUA------CUUGG--GCAUAUUACGCAUACGACAGGUAUGC------------AAUGACAUUGAUCAACUGCUG -(((((((..(((((((((((.((....(((((...))))------).)).--))))))...((((((.....))))))------------...))))))))))))...... ( -29.30) >DroSec_CAF1 38517 91 + 1 -UUGGUCAACGUGUCGUAUGCGCACUUUGUAUAUCGUAUA------CUUGG--GCAUAUUACGCAUACGACAGGUAUGC------------AAUGACAUUGAUCAACUGCUG -(((((((..(((((((((((.((....(((((...))))------).)).--))))))...((((((.....))))))------------...))))))))))))...... ( -29.30) >DroSim_CAF1 37580 91 + 1 -UUGGCCAACGUGUCGUAUGCGCACUUUGUAUAUUGUAUA------CUUGG--GCAUAUUACGCAUACGACAGGUAUGC------------AAUGACAUUGAUGAACUGCUG -...((..((.((((((((((((.((..(((((...))))------)..))--)).......)))))))))).))..))------------..................... ( -24.61) >DroEre_CAF1 38459 76 + 1 -UUGGUCAACAUGUCGUAUGCGCACUU---------------------UGG--GCAUAUUACGCAUACGACAGGUAUGC------------AAUGACAUUGAUCAACUGCUG -(((((((..(((((((((((.(....---------------------.).--))))))...((((((.....))))))------------...))))))))))))...... ( -26.70) >DroYak_CAF1 41239 91 + 1 -UUCGUCAACGCGUCGUAUGCGCACUUUGUAUAGUGUAUA------CUUGG--GCAUAUUACGCAUACGACAGGUAUGC------------AAUGACAUUGAUCAACUGCUG -...((((....((.((((((.((....(((((...))))------).)).--)))))).))((((((.....))))))------------..))))............... ( -24.80) >DroPer_CAF1 46708 110 + 1 GUGGGGCAACGUGUUGCAUACGU--UAUAUAUAUUAGAUAGAACCCCCAGAGCACACAUUACGCAUACGAGAGGUAUGCCCCCUGUUGUUUAGUUCAAUUAGUCAACUGCAG .(((((.(((((((...))))))--)...(((.....)))....)))))..(((........((((((.....)))))).....((((.(((((...))))).))))))).. ( -24.10) >consensus _UUGGUCAACGUGUCGUAUGCGCACUUUGUAUAUUGUAUA______CUUGG__GCAUAUUACGCAUACGACAGGUAUGC____________AAUGACAUUGAUCAACUGCUG ........((.(((((((((((.......................................))))))))))).))..................................... (-14.42 = -14.95 + 0.53)


| Location | 6,421,559 – 6,421,650 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.21 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -16.89 |
| Energy contribution | -16.83 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6421559 91 - 22224390 CAGCAGUUGAUCAAUGUCAUU------------GCAUACCUGUCGUAUGCGUAAUAUGC--CCAAG------UAUAUAAUAUACAAAGUGCGCAUACGACACGUUGACCAA- ..(((((.(((....))))))------------))..((.(((((((((((((((((((--....)------)))))...........))))))))))))).)).......- ( -28.30) >DroSec_CAF1 38517 91 - 1 CAGCAGUUGAUCAAUGUCAUU------------GCAUACCUGUCGUAUGCGUAAUAUGC--CCAAG------UAUACGAUAUACAAAGUGCGCAUACGACACGUUGACCAA- ..(((((.(((....))))))------------))..((.(((((((((((((.(((((--....)------))))............))))))))))))).)).......- ( -27.12) >DroSim_CAF1 37580 91 - 1 CAGCAGUUCAUCAAUGUCAUU------------GCAUACCUGUCGUAUGCGUAAUAUGC--CCAAG------UAUACAAUAUACAAAGUGCGCAUACGACACGUUGGCCAA- ..(((((.((....))..)))------------))..((.(((((((((((((.(((((--....)------))))............))))))))))))).)).......- ( -24.92) >DroEre_CAF1 38459 76 - 1 CAGCAGUUGAUCAAUGUCAUU------------GCAUACCUGUCGUAUGCGUAAUAUGC--CCA---------------------AAGUGCGCAUACGACAUGUUGACCAA- ..(((((.(((....))))))------------))..((.(((((((((((((...(..--...---------------------.).))))))))))))).)).......- ( -26.00) >DroYak_CAF1 41239 91 - 1 CAGCAGUUGAUCAAUGUCAUU------------GCAUACCUGUCGUAUGCGUAAUAUGC--CCAAG------UAUACACUAUACAAAGUGCGCAUACGACGCGUUGACGAA- ..(((((.(((....))))))------------))..((.(((((((((((...(((((--....)------))))((((......))))))))))))))).)).......- ( -27.90) >DroPer_CAF1 46708 110 - 1 CUGCAGUUGACUAAUUGAACUAAACAACAGGGGGCAUACCUCUCGUAUGCGUAAUGUGUGCUCUGGGGGUUCUAUCUAAUAUAUAUA--ACGUAUGCAACACGUUGCCCCAC (((..(((..............)))..)))((((((.((.....((((((((.(((((((...((((.......)))).))))))).--)))))))).....)))))))).. ( -29.34) >consensus CAGCAGUUGAUCAAUGUCAUU____________GCAUACCUGUCGUAUGCGUAAUAUGC__CCAAG______UAUACAAUAUACAAAGUGCGCAUACGACACGUUGACCAA_ ..................................((.((.(((((((((((((...................................))))))))))))).))))...... (-16.89 = -16.83 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:33 2006