
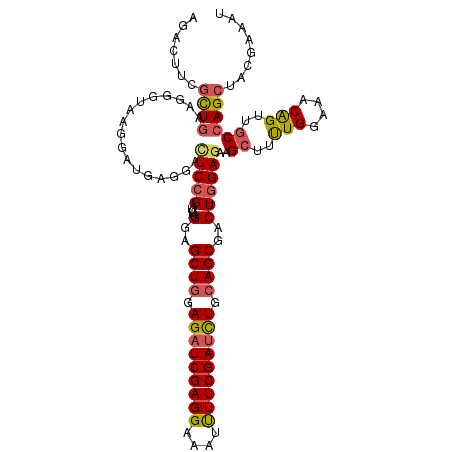
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,405,922 – 6,406,074 |
| Length | 152 |
| Max. P | 0.950070 |

| Location | 6,405,922 – 6,406,034 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 89.46 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -30.40 |
| Energy contribution | -30.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6405922 112 + 22224390 AGACAUCGCUGAAGGGCAAGGAUGAGGACUCCGAUUUGGAGCUGGAGAUCGAGGAAAUUUUCGAUUUGCAGCGACUGGAGAAGCUUUUGGAAACAGUUGCCAGCUACGAAAU .....(((..(...(((((....((((.(((((....(..((((.(((((((((....))))))))).))))..))))))...))))((....)).)))))..)..)))... ( -36.60) >DroSec_CAF1 31328 112 + 1 AGACUUCGUUGCAGGGUAAGGAUGAGGACUCCGAUUUGGAGCUGGAGAUCGAGGAAAUUUUCGAUCUGCAGCGACUGGAGAAGCUAUUGGAAACAGUUGCCAGCUACGAAAU ....(((((.((..(((((.........(((((....(..((((.(((((((((....))))))))).))))..))))))......(((....)))))))).)).))))).. ( -41.10) >DroSim_CAF1 30471 112 + 1 AGACUUCGCUGAAGGGUAAGGAUGAGGAUUCCGAUUUGGAGCUGGAGAUCGAGGACAUUUUCGAUCUGCAGCGACUGGAGAAGCUAUUGGAAACAGUUGCCAGCUACGAAAU .......((((........(((.......)))..((..(.((((.(((((((((....))))))))).))))..)..))...((.((((....)))).))))))........ ( -36.90) >DroEre_CAF1 31481 112 + 1 AGACUUCGCUGAAGGGUAAGGAUGAGGACUCCGAUUUGGAGCUGGAGAUCGAGGAAAUUUUCGAUCUGCAGCGACUGGAGAAGCUUUUGGAAACGGUUGCAAGCUACGAAAU ...(((((((.........)).))))).(((((....(..((((.(((((((((....))))))))).))))..))))))..((..(((....)))..))............ ( -35.80) >DroYak_CAF1 34699 112 + 1 AGACUUCGCUGAAGGGUAAGGAUGAGGACUCAGAUUUGGAGCUAGAGAUCGAGGAAAUUUUCGAUCUUCAGCGACUGGAGAAGCUUUUGGAAACAGUUGCCAGCUACGAACU .......((((..((((..........))))...((..(.(((..(((((((((....)))))))))..)))..)..))...((..(((....)))..))))))........ ( -32.40) >DroAna_CAF1 41526 112 + 1 AGCCAUCCCUGAAGGGCAGGGAGGACGACUCCGACCUGGAGCUGGAGAUCGAGGAGAUCUUCGACUUGCAGCGCCUGGAACAGUUGCUGGAAACGGUGUCCAGCUACGAGAU (((..((((((.....))))))((((((((....((.((.((((.((.((((((....)))))).)).)))).)).))...))))((((....)))))))).)))....... ( -47.60) >consensus AGACUUCGCUGAAGGGUAAGGAUGAGGACUCCGAUUUGGAGCUGGAGAUCGAGGAAAUUUUCGAUCUGCAGCGACUGGAGAAGCUUUUGGAAACAGUUGCCAGCUACGAAAU .......((((.................(((((....(..((((.(((((((((....))))))))).))))..))))))..((..(((....)))..))))))........ (-30.40 = -30.60 + 0.20)


| Location | 6,405,954 – 6,406,074 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.52 |
| Mean single sequence MFE | -43.40 |
| Consensus MFE | -25.86 |
| Energy contribution | -26.07 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6405954 120 + 22224390 GAUUUGGAGCUGGAGAUCGAGGAAAUUUUCGAUUUGCAGCGACUGGAGAAGCUUUUGGAAACAGUUGCCAGCUACGAAAUGCGUCGCCGCAUUCGUGCACAAAUGCGUCUUAUACGCAAG ..((..(.((((.(((((((((....))))))))).))))..)..))...((..(((....)))..))..((.(((((.((((....))))))))))).....(((((.....))))).. ( -42.80) >DroPse_CAF1 37981 116 + 1 ----UCGAUCUGGAGAUUGAGGAGAUCUUCGAUCUGCAGGUACUCGAACAGCUCCUGGAGACAGUGAGCAGCUACGAGAUGCGUCGCCGGAUACGCGCCCAGAUGCGGCUCAUCAGAAAG ----((((((((.(((((((((....))))))))).))))...))))...(((((((....))).))))......(((.((((((((((....)).))...)))))).)))......... ( -47.50) >DroWil_CAF1 32757 116 + 1 ----UCGAUUUGGAAAUCGAAGAGAUAUUCGAUCUGCAGAUCUUGGAACAACUUUUAGAGACCGUAACCAGUUACGAGUUGCGUCGUCGUAUACGAGCUCAGAUGCGUUUGAUCAGAAAG ----..((((((.(.((((((......)))))).).)))))).........(((((.((((((((((....))))).)))(((((.(((....))).....)))))......)).))))) ( -30.20) >DroMoj_CAF1 43037 112 + 1 --------GCUGGAGAUUGAGGAGAUCUUCGAUCUGCAGACGCUGGAGCAGCUGCUGGAGACGGUGAGCAGCUAUGAGCUGCGUCGACGCAUUCGCGCCCAGAUACGUCUGAUUAAGAAG --------.....(((((((((....))))))))).((((((((((.((((((((((....)....)))))))..(((.((((....)))))))..))))))...))))))......... ( -45.50) >DroAna_CAF1 41558 120 + 1 GACCUGGAGCUGGAGAUCGAGGAGAUCUUCGACUUGCAGCGCCUGGAACAGUUGCUGGAAACGGUGUCCAGCUACGAGAUGCGCCGCAGGAUCCGCGCCCAAAUGCGCCUCAUCCGGAAG ...(((((...(((..((((((....))))))(((((.(((((((((......((((....)))).))))).........)))).))))).)))((((......))))....)))))... ( -46.90) >DroPer_CAF1 37999 116 + 1 ----UCGAUCUGGAGAUUGAGGAGAUCUUCGAUCUGCAGGUACUCGAACAGCUCCUGGAGACAGUGAGCAGCUACGAGAUGCGUCGCCGGAUACGCGCCCAGAUGCGGCUCAUCAGAAAG ----((((((((.(((((((((....))))))))).))))...))))...(((((((....))).))))......(((.((((((((((....)).))...)))))).)))......... ( -47.50) >consensus ____UCGAGCUGGAGAUCGAGGAGAUCUUCGAUCUGCAGAUACUGGAACAGCUCCUGGAGACAGUGACCAGCUACGAGAUGCGUCGCCGGAUACGCGCCCAGAUGCGUCUCAUCAGAAAG ........((((.(((((((((....))))))))).)))).........((((.(((....))).....))))..(((.((((((((((....)).))...)))))).)))......... (-25.86 = -26.07 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:30 2006