| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,404,626 – 6,404,774 |
| Length | 148 |
| Max. P | 0.801641 |

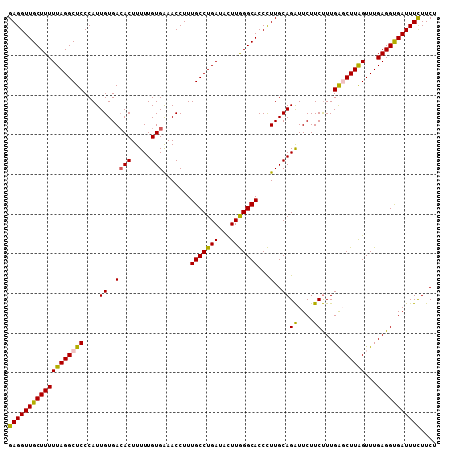
| Location | 6,404,626 – 6,404,734 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.14 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -23.92 |
| Energy contribution | -23.55 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6404626 108 - 22224390 GAGGUUGCUUUUUAGGCUCCCAUUGUGACACUUUUUGUGAAACCUUUGCCUGAUACUUGGGCACCCUUGCAGAUUCUUCUUUGACCUUGGUUUGAGGUGAUUUCUUCU ((((.(((((.((((((.......((..(((.....)))..))....)))))).....))))).))))(.(((.((.((((.(((....))).)))).))..))).). ( -26.60) >DroSec_CAF1 30044 108 - 1 GAGGUUGCUUUUUAGGCUCCCAUUGUGACACUUUUUGUGAAACCUUUGCCUGAUACUUGGGCACCCUUGCAGAUUCUUCUUUGGGCUUAGUUUGAGGUGAUUUCUUCU (((((..((((((((((((....((..((((.....))).......((((..(...)..))))...)..))((....))...))))))))...))))..))))).... ( -27.50) >DroSim_CAF1 29175 108 - 1 GAGGUUGCUUUUUAGGCUCCCAUUGUGACACUUUUUGUGAAACCUUUGCCUGAUACUUGGGCACCCUUGCAGAUUCUUCUUUGAUCUUAGUUUGAGGCGAUUUCUUCU ((((((((...((((((.......((..(((.....)))..))....))))))..((..(((.......((((......))))......)))..)))))))))).... ( -25.12) >DroYak_CAF1 33391 108 - 1 GAGGUUGCUUUUUAGGCUCCCAUUGUGAAACUUUUUGUGGAACCUUUGCCUGAUAUUUAGGCACCUUUGCAGGUUCUUCCUUGAGCUUAGUUUGAGGUGAUUUUUUCU (((((((((.....)))..((((.............)))))))))).((((((...))))))(((((.((((((((......)))))).))..))))).......... ( -28.22) >consensus GAGGUUGCUUUUUAGGCUCCCAUUGUGACACUUUUUGUGAAACCUUUGCCUGAUACUUGGGCACCCUUGCAGAUUCUUCUUUGAGCUUAGUUUGAGGUGAUUUCUUCU (((((((((((((((((((....((..((((.....))).......(((((((...)))))))...)..))((....))...))))))))...))))))))))).... (-23.92 = -23.55 + -0.37)


| Location | 6,404,660 – 6,404,774 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 93.86 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -22.22 |
| Energy contribution | -23.47 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6404660 114 + 22224390 AUCUGCAAGGGUGCCCAAGUAUCAGGCAAAGGUUUCACAAAAAGUGUCACAAUGGGAGCCUAAAAAGCAACCUCAAAGGGAGCCAAAGGUAACUCAAAAGGAAACCCCAUUGGA ...(((...(((((....)))))..))).......(((.....)))...(((((((.((((.....((..((.....))..))...)))).........(....)))))))).. ( -28.20) >DroSec_CAF1 30078 114 + 1 AUCUGCAAGGGUGCCCAAGUAUCAGGCAAAGGUUUCACAAAAAGUGUCACAAUGGGAGCCUAAAAAGCAACCUCAAAGGGAGCCAAAGGUGACUCAAAAGGAAACCCCAUGGGA ...(((...(((((....)))))..)))..((((((.........(((((....((..(((....((....))....)))..))....))))).......))))))........ ( -28.39) >DroSim_CAF1 29209 114 + 1 AUCUGCAAGGGUGCCCAAGUAUCAGGCAAAGGUUUCACAAAAAGUGUCACAAUGGGAGCCUAAAAAGCAACCUCAAAGGGAGCCGAAGGUGACUCAAAAGGAAACCCCAUGGGA ...(((...(((((....)))))..)))..((((((.........(((((..(.((..(((....((....))....)))..)).)..))))).......))))))........ ( -28.49) >DroYak_CAF1 33425 114 + 1 ACCUGCAAAGGUGCCUAAAUAUCAGGCAAAGGUUCCACAAAAAGUUUCACAAUGGGAGCCUAAAAAGCAACCUCUACGGGAGCCAAAGGUGUCACAAAAAGAAACCCCAUGGGA .((((...((((((((.......))))..(((((((.((.............)))))))))........))))...))))..(((..(((.((.......)).)))...))).. ( -29.62) >consensus AUCUGCAAGGGUGCCCAAGUAUCAGGCAAAGGUUUCACAAAAAGUGUCACAAUGGGAGCCUAAAAAGCAACCUCAAAGGGAGCCAAAGGUGACUCAAAAGGAAACCCCAUGGGA ...(((...(((((....)))))..)))..((((((.........(((((....((..(((....((....))....)))..))....))))).......))))))........ (-22.22 = -23.47 + 1.25)

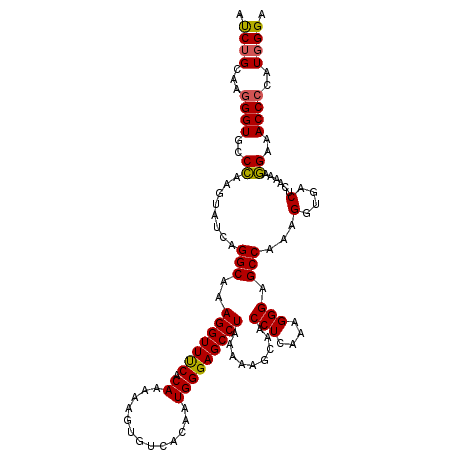
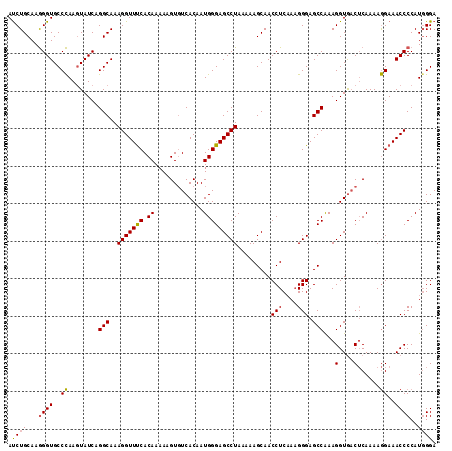
| Location | 6,404,660 – 6,404,774 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 93.86 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -27.00 |
| Energy contribution | -26.75 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6404660 114 - 22224390 UCCAAUGGGGUUUCCUUUUGAGUUACCUUUGGCUCCCUUUGAGGUUGCUUUUUAGGCUCCCAUUGUGACACUUUUUGUGAAACCUUUGCCUGAUACUUGGGCACCCUUGCAGAU ((((((((((...(((...((((.(((((.((...))...))))).))))...))).)))))))).)).....((((..(......((((..(...)..))))...)..)))). ( -33.90) >DroSec_CAF1 30078 114 - 1 UCCCAUGGGGUUUCCUUUUGAGUCACCUUUGGCUCCCUUUGAGGUUGCUUUUUAGGCUCCCAUUGUGACACUUUUUGUGAAACCUUUGCCUGAUACUUGGGCACCCUUGCAGAU .....(((((...(((...((((.(((((.((...))...))))).))))...))).)))))(((..((((.....))).......((((..(...)..))))...)..))).. ( -30.60) >DroSim_CAF1 29209 114 - 1 UCCCAUGGGGUUUCCUUUUGAGUCACCUUCGGCUCCCUUUGAGGUUGCUUUUUAGGCUCCCAUUGUGACACUUUUUGUGAAACCUUUGCCUGAUACUUGGGCACCCUUGCAGAU .....(((((...(((...((((.(((((.((...))...))))).))))...))).)))))(((..((((.....))).......((((..(...)..))))...)..))).. ( -29.20) >DroYak_CAF1 33425 114 - 1 UCCCAUGGGGUUUCUUUUUGUGACACCUUUGGCUCCCGUAGAGGUUGCUUUUUAGGCUCCCAUUGUGAAACUUUUUGUGGAACCUUUGCCUGAUAUUUAGGCACCUUUGCAGGU ..(((.((((((.(.....).))).))).)))..((.((((((((.((((.(((((((((((..(.......)..)).)))......)))))).....)))))))))))).)). ( -33.00) >consensus UCCCAUGGGGUUUCCUUUUGAGUCACCUUUGGCUCCCUUUGAGGUUGCUUUUUAGGCUCCCAUUGUGACACUUUUUGUGAAACCUUUGCCUGAUACUUGGGCACCCUUGCAGAU (((.((((((...(((...((((.(((((.((...))...))))).))))...))).)))))).).)).....((((..(......(((((((...)))))))...)..)))). (-27.00 = -26.75 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:28 2006