
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,357,225 – 6,357,410 |
| Length | 185 |
| Max. P | 0.915127 |

| Location | 6,357,225 – 6,357,341 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.97 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -13.21 |
| Energy contribution | -12.65 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6357225 116 + 22224390 GUAGGGGAAUCGAGAG-AACACGA-AACGACAGGAUUUCGUGAAGAGAUAUUUCUGCCGAAAGGAGAAGAAGUAGGAGAAUGC-CCCA-UGACUAAGCCAAUAACCGAAAGGUUAUUUAA ...((((..((....)-).(((((-((........)))))))......(((((((.((....))...)))))))........)-))).-..........(((((((....)))))))... ( -36.90) >DroPse_CAF1 36008 115 + 1 A-AACGGAAUGGAAUGAAACAAGGAAAUG--AGGAAUAUGCGACUGGCA--CUAUACUGAAGGGAUGGGAUGGGGAUGGAUGCGCUCACGGACUAAGGCAAUAACAGAAAGGUUAUUUAA .-..(((.((((.......((......))--.......(((.....)))--)))).)))......(((..(((((.((....))))).))..)))....((((((......))))))... ( -13.10) >DroSec_CAF1 41243 111 + 1 GUAGGGGAAUGG--CG-AAUACGG-AACGACAGGAUUUCGUGAGGGGAUAUUUCUGCCGAAAGGAGAAGAA---GGAUAAUGC-CCCA-UGACUAAGCCAAUAACCAAAAGGUUAUUUAA .........(((--(.-..(((((-((........))))))).((((...(((((.((....))...))))---).......)-))).-.......))))((((((....)))))).... ( -32.20) >DroEre_CAF1 39103 111 + 1 GUAGAGGAAUGG--AG-AACAUGG-AACGACAGGAUUUCGUGAAGAGAUAUUUCUGCCCAAAAGGGAAGAA---GUAGAAUAC-CUCA-UGACUAAGCCAAUAACCAAAAGGUUAUUUAA ...((((.....--..-..(((((-((........)))))))......(((((((.(((....))).))))---))).....)-))).-..........(((((((....)))))))... ( -27.60) >consensus GUAGGGGAAUGG__AG_AACACGG_AACGACAGGAUUUCGUGAAGAGAUAUUUCUGCCGAAAGGAGAAGAA___GGAGAAUGC_CCCA_UGACUAAGCCAAUAACCAAAAGGUUAUUUAA ...(((.............(((((...(....)....))))).........((((.((....))))))................)))............(((((((....)))))))... (-13.21 = -12.65 + -0.56)



| Location | 6,357,263 – 6,357,378 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.34 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -12.05 |
| Energy contribution | -11.55 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.45 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6357263 115 + 22224390 UGAAGAGAUAUUUCUGCCGAAAGGAGAAGAAGUAGGAGAAUGC-CCCA-UGACUAAGCCAAUAACCGAAAGGUUAUUUAAUAUUUGGC---AGUGGCGUGGCCUUAAAAAAAUACGAGCA ........(((((((.((....))...)))))))((...((((-(...-((.(((((..(((((((....))))))).....))))))---)..)))))..))................. ( -33.00) >DroPse_CAF1 36045 97 + 1 CGACUGGCA--CUAUACUGAAGGGAUGGGAUGGGGAUGGAUGCGCUCACGGACUAAGGCAAUAACAGAAAGGUUAUUUAAUAUUUGACUACAGUUGGCU--------------------- ((((((((.--((...((....)).(((..(((((.((....))))).))..)))))))((((((......)))))).............))))))...--------------------- ( -16.10) >DroSec_CAF1 41279 112 + 1 UGAGGGGAUAUUUCUGCCGAAAGGAGAAGAA---GGAUAAUGC-CCCA-UGACUAAGCCAAUAACCAAAAGGUUAUUUAAUAUUUGGC---AGUGGCGUGGCCUUAAAAAAAUACGAGAA (((((((...(((((.((....))...))))---).......)-)(((-((.(((.((((((((((....))))))........))))---..))))))))))))).............. ( -30.70) >DroEre_CAF1 39139 108 + 1 UGAAGAGAUAUUUCUGCCCAAAAGGGAAGAA---GUAGAAUAC-CUCA-UGACUAAGCCAAUAACCAAAAGGUUAUUUAAUAUUUGGC---AGUGGCGUGGCCUUAAAAAAUAAG----G ........(((((((.(((....))).))))---)))......-.(((-((.(((.((((((((((....))))))........))))---..))))))))(((((.....))))----) ( -27.80) >consensus UGAAGAGAUAUUUCUGCCGAAAGGAGAAGAA___GGAGAAUGC_CCCA_UGACUAAGCCAAUAACCAAAAGGUUAUUUAAUAUUUGGC___AGUGGCGUGGCCUUAAAAAAAUAC____A ...........((((.((....)))))).......................(((..((((((((((....))))))........))))...))).......................... (-12.05 = -11.55 + -0.50)

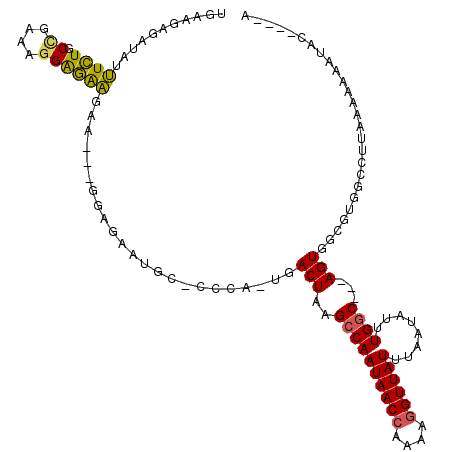

| Location | 6,357,303 – 6,357,410 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -17.65 |
| Energy contribution | -17.43 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6357303 107 + 22224390 UGCCCCAUGACUAAGCCAAUAACCGAAAGGUUAUUUAAUAUUUGGCAGUGGCGUGGCCUUAAAAAAAUACGAGCAAAAACACCCUUUCUUGGC--------UCCACUGCUCCACA ....(((((.(((.((((((((((....))))))........))))..))))))))..............(((((.......((......)).--------.....))))).... ( -27.72) >DroSec_CAF1 41316 115 + 1 UGCCCCAUGACUAAGCCAAUAACCAAAAGGUUAUUUAAUAUUUGGCAGUGGCGUGGCCUUAAAAAAAUACGAGAAAAAACACCCUUUCUUGGCUCUACGGCUCCACUGGUCCACA .(((...((.(((((..(((((((....))))))).....)))))))..)))((((((...........(((((((........)))))))(((....)))......)).)))). ( -29.30) >DroEre_CAF1 39176 96 + 1 UACCUCAUGACUAAGCCAAUAACCAAAAGGUUAUUUAAUAUUUGGCAGUGGCGUGGCCUUAAAAAAUAAG----GAAAACACCCUUUUUUUG---------------GCUCCACA ..............((((((((((....))))))........)))).((((...((((....((((.(((----(.......)))).)))))---------------))))))). ( -23.30) >consensus UGCCCCAUGACUAAGCCAAUAACCAAAAGGUUAUUUAAUAUUUGGCAGUGGCGUGGCCUUAAAAAAAUACGAG_AAAAACACCCUUUCUUGGC________UCCACUGCUCCACA ....(((((.(((.((((((((((....))))))........))))..))))))))........................................................... (-17.65 = -17.43 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:14 2006