
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,350,618 – 6,350,766 |
| Length | 148 |
| Max. P | 0.930110 |

| Location | 6,350,618 – 6,350,728 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -15.45 |
| Energy contribution | -16.57 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6350618 110 + 22224390 AUUAUAAUUAUACUAAUCAAUUCCACAACUUAGUGCACCCGAGCGCAAAGUUGUACAAUUUUGUGAGAGCCCUGAGGGACUUUUAGUCCUUUUUUGUU-----UUUUUUUUUUGC ........................(((((((.((((......)))).))))))).(((......((((((...(((((((.....)))))))...)))-----))).....))). ( -26.30) >DroSec_CAF1 34619 115 + 1 AUUAUAAUUAUACUAAUCAAUUCCACAACUUAGUGCACCCGAGCGCAAAGUUGUACAAUUUUGUGAGAGCCCUGAGGGACUUUUAGUCCUUUUUCUUUUUUUCUUUUUUUUUUGU ........................(((((((.((((......)))).)))))))((((......(((((....(((((((.....)))))))........)))))......)))) ( -24.40) >DroEre_CAF1 32799 93 + 1 AUUAUAAUUUUACUAAUCAAUUCCAAAACUUAGGGCACCCGAGCGCAAAGUUGUACAAUUUUGUGAGAGCCUUG-GGUAC-UUUAGUCCCUUAUU-------------------- .............................((((((.((((((((((((((((....))))))))).....))))-))).)-))))).........-------------------- ( -20.10) >consensus AUUAUAAUUAUACUAAUCAAUUCCACAACUUAGUGCACCCGAGCGCAAAGUUGUACAAUUUUGUGAGAGCCCUGAGGGACUUUUAGUCCUUUUUU_UU_____UUUUUUUUUUG_ ............((...(((....(((((((.((((......)))).)))))))......)))..))......(((((((.....)))))))....................... (-15.45 = -16.57 + 1.11)

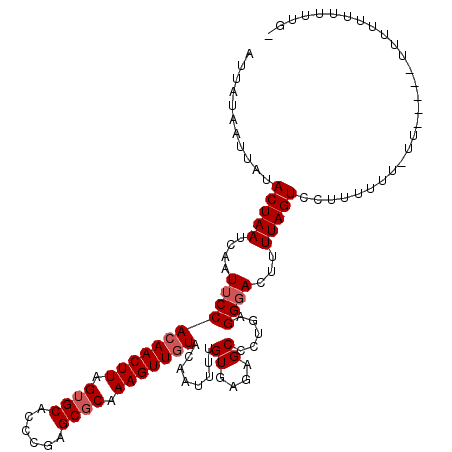
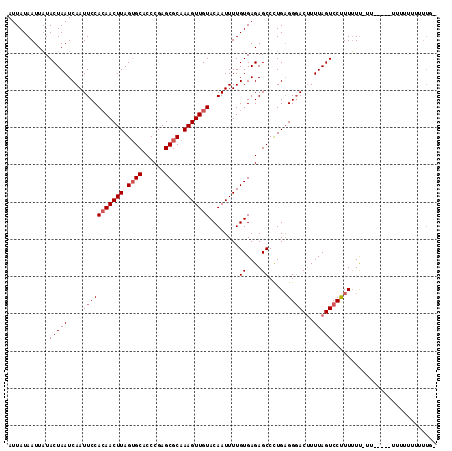
| Location | 6,350,618 – 6,350,728 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -18.77 |
| Energy contribution | -19.77 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6350618 110 - 22224390 GCAAAAAAAAAA-----AACAAAAAAGGACUAAAAGUCCCUCAGGGCUCUCACAAAAUUGUACAACUUUGCGCUCGGGUGCACUAAGUUGUGGAAUUGAUUAGUAUAAUUAUAAU ............-----...........(((((.(((((....))))).......((((.(((((((((((((....)))))..)))))))).))))..)))))........... ( -24.80) >DroSec_CAF1 34619 115 - 1 ACAAAAAAAAAAGAAAAAAAGAAAAAGGACUAAAAGUCCCUCAGGGCUCUCACAAAAUUGUACAACUUUGCGCUCGGGUGCACUAAGUUGUGGAAUUGAUUAGUAUAAUUAUAAU ............................(((((.(((((....))))).......((((.(((((((((((((....)))))..)))))))).))))..)))))........... ( -24.80) >DroEre_CAF1 32799 93 - 1 --------------------AAUAAGGGACUAAA-GUACC-CAAGGCUCUCACAAAAUUGUACAACUUUGCGCUCGGGUGCCCUAAGUUUUGGAAUUGAUUAGUAAAAUUAUAAU --------------------.((((...(((((.-(((((-(.((((......................)).)).))))))((........))......)))))....))))... ( -17.35) >consensus _CAAAAAAAAAA_____AA_AAAAAAGGACUAAAAGUCCCUCAGGGCUCUCACAAAAUUGUACAACUUUGCGCUCGGGUGCACUAAGUUGUGGAAUUGAUUAGUAUAAUUAUAAU ............................(((((..((((....))))........((((.((((((((.((((....))))...)))))))).))))..)))))........... (-18.77 = -19.77 + 1.00)

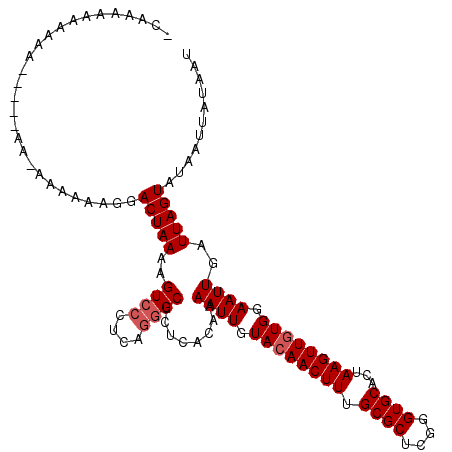
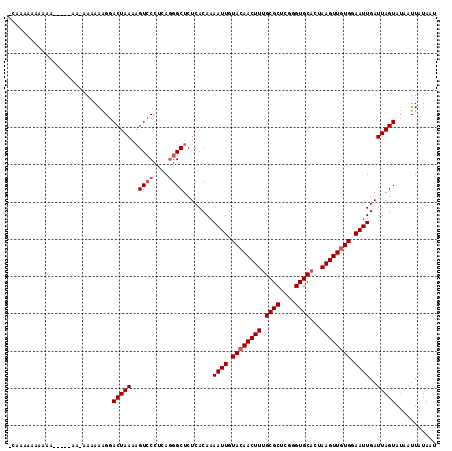
| Location | 6,350,653 – 6,350,766 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -23.99 |
| Energy contribution | -24.43 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6350653 113 + 22224390 CACCCGAGCGCAAAGUUGUACAAUUUUGUGAGAGCCCUGAGGGACUUUUAGUCCUUUUUUGUU-----UUUUUUUUUUGCCAAGUGGAAAGGGUGGAAA--AGCAGGCACUGACUGAUUU (((((...(((((((((....)))))))))(((((...(((((((.....)))))))...)))-----))...((((..(...)..)))))))))....--..(((.......))).... ( -31.50) >DroSec_CAF1 34654 118 + 1 CACCCGAGCGCAAAGUUGUACAAUUUUGUGAGAGCCCUGAGGGACUUUUAGUCCUUUUUCUUUUUUUCUUUUUUUUUUGUGAAGUGGAAAGGGUGGAAA--AGCAGGCACUGACUGAUUU (((((...(((((((((....)))))))))((((....(((((((.....)))))))..)))).(((((.((((......)))).))))))))))....--..(((.......))).... ( -31.00) >DroEre_CAF1 32834 96 + 1 CACCCGAGCGCAAAGUUGUACAAUUUUGUGAGAGCCUUG-GGUAC-UUUAGUCCCUUAUU--------------------CAAAUUG--AGGGUGGAAGGCACCAGGCACUGACUGAUUU ........(((((((((....)))))))))((.((((.(-(...(-(((...((((((..--------------------.....))--))))..))))...)))))).))......... ( -29.80) >consensus CACCCGAGCGCAAAGUUGUACAAUUUUGUGAGAGCCCUGAGGGACUUUUAGUCCUUUUUU_UU_____UUUUUUUUUUG_CAAGUGGAAAGGGUGGAAA__AGCAGGCACUGACUGAUUU (((((...(((((((((....)))))))))........(((((((.....))))))).................................)))))........(((.......))).... (-23.99 = -24.43 + 0.45)

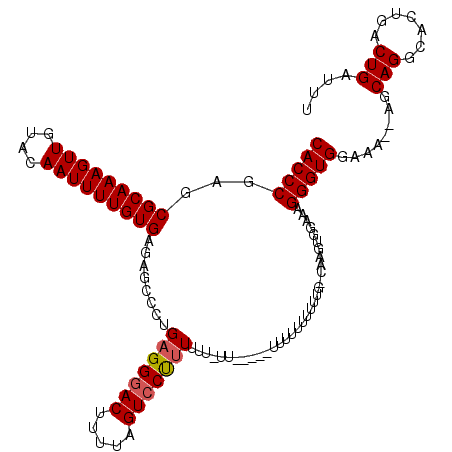
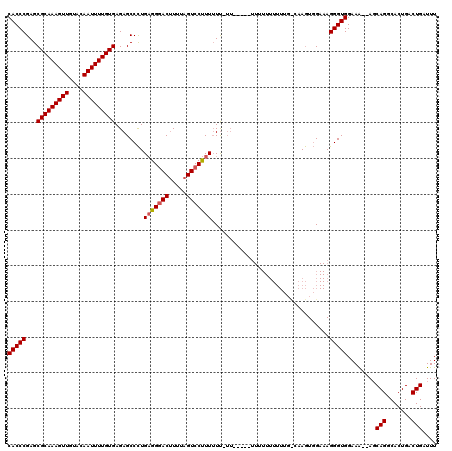
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:09 2006