
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,350,020 – 6,350,161 |
| Length | 141 |
| Max. P | 0.815559 |

| Location | 6,350,020 – 6,350,121 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.54 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -15.49 |
| Energy contribution | -15.60 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6350020 101 + 22224390 GUAAUACCUUCGCUGCCAGCAGCCCUUUUUCCACCA------CCCAAAGCAUGGA-------------AAACCCCAGCGCUCGGAAACCACCAAGCCAAAUGGCAGCCCCAUGCUAACCC ...........(((((((((......(((((((...------.........))))-------------))).....))(((.((......)).)))....)))))))............. ( -23.40) >DroSec_CAF1 34052 100 + 1 GUAAUACCUCCGCUGCCAGCAGCCCUU-UUCCUCCA------CCCGGUGCCUGGA-------------AAACCGCAACGCUCGGAAACCACCAAGCCAAAUGGCAGCCCCAUGCUAUCCC ...........(((((((...((..((-((((..((------(...)))...)))-------------)))..))...(((.((......)).)))....)))))))............. ( -26.80) >DroEre_CAF1 32069 119 + 1 GUAAUACUGCCGCUGGCAGCAGCCCUUUUCCGUACAGUAUACCCCAGACCCUGGAAGCACCCAUCUCAAAGCCCCAACCCUCGGAAACCACCAAGCCAAAUGGCAGCC-CAAGCUAUCCC ......(((((((((....))))...((((((....((.....((((...))))..((............))....))...))))))..............)))))..-........... ( -22.00) >consensus GUAAUACCUCCGCUGCCAGCAGCCCUUUUUCCUCCA______CCCAGAGCCUGGA_____________AAACCCCAACGCUCGGAAACCACCAAGCCAAAUGGCAGCCCCAUGCUAUCCC ...........(((((((...((....................(((.....)))............................((......))..))....)))))))............. (-15.49 = -15.60 + 0.11)

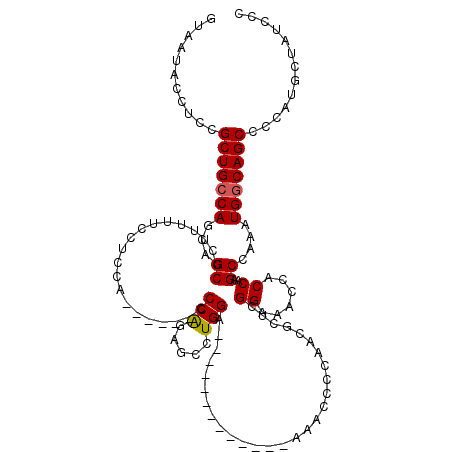

| Location | 6,350,056 – 6,350,161 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.87 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -18.67 |
| Energy contribution | -19.23 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6350056 105 + 22224390 --CCCAAAGCAUGGA-------------AAACCCCAGCGCUCGGAAACCACCAAGCCAAAUGGCAGCCCCAUGCUAACCCAAACACAAGAGGACUCCUUGACUUAACCCACCAGGUGCUG --.(((.....))).-------------......((((((..((..........(((....)))(((.....)))..........((((.......))))..........))..)))))) ( -22.60) >DroSec_CAF1 34087 105 + 1 --CCCGGUGCCUGGA-------------AAACCGCAACGCUCGGAAACCACCAAGCCAAAUGGCAGCCCCAUGCUAUCCCAAACCCGAGAGAACUCCUUGACAUAACCCACCAGGUGCUG --..(((..(((((.-------------.........(.(((((..........(((....)))(((.....))).........))))).).......((........)))))))..))) ( -29.40) >DroEre_CAF1 32109 119 + 1 ACCCCAGACCCUGGAAGCACCCAUCUCAAAGCCCCAACCCUCGGAAACCACCAAGCCAAAUGGCAGCC-CAAGCUAUCCCAAACCGGAGAGAACUCCUUGACAUAGCCCACCAGGUGCUG ....(((..(((((..((........................((......))..(((....))).)).-...(((((..(((...((((....)))))))..)))))...)))))..))) ( -29.60) >consensus __CCCAGAGCCUGGA_____________AAACCCCAACGCUCGGAAACCACCAAGCCAAAUGGCAGCCCCAUGCUAUCCCAAACCCGAGAGAACUCCUUGACAUAACCCACCAGGUGCUG ....(((..(((((............................((..........(((....)))(((.....)))...)).....((((.(....)))))..........)))))..))) (-18.67 = -19.23 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:07 2006