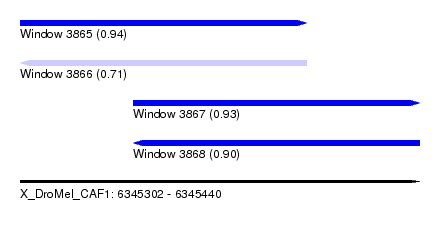
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,345,302 – 6,345,440 |
| Length | 138 |
| Max. P | 0.935377 |

| Location | 6,345,302 – 6,345,401 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 93.37 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -35.50 |
| Energy contribution | -36.25 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6345302 99 + 22224390 UUUUGAAUGAGUCUCCACGGAAUAAGCGCGUUCGCAUCGCUGACUAUCUGAGCAGCGGCAGAGCUCCAACCGGAGCUCCAAUUGGAGGUCAGUGGUGG--------C ....(((((.((.((....))....)).))))).(((((((((((.((..(((....)).(((((((....)))))))...)..))))))))))))).--------. ( -39.10) >DroSec_CAF1 29684 99 + 1 UUUUGAAUGAGUCUCCACGGAAUAAGCGCGUCCGCAUCGCUGACUAUCUGAGCAGCGGCAGAGCUCCAAGUGGAGCUCCAAUUGGAGGUCAGUGGUGG--------C ..........(((.((((.......((((....))..(((((.((.....))))))))).((.((((((.(((....))).)))))).)).)))).))--------) ( -38.00) >DroEre_CAF1 27572 99 + 1 UUUUGAAUGAGUCUCCACGGAAUAAGCGCGUUCGCAUCGCUGACUAUCUGAGCAGCGGCAGAGCUCCAAUUGGAGCUCCAAUUGCAGGUCAGUGGUGG--------C ....(((((.((.((....))....)).))))).(((((((((((......((....)).(((((((....)))))))........))))))))))).--------. ( -36.60) >DroYak_CAF1 26305 107 + 1 UUUUGAAUGAGUCUCCACGGAAUAAGCGCGUUCGCAUCGCUGACUAUCUGAGCAGCGGCAGAGCUCCAACUGGAGCUCCAAUUGGAGGUCAGUGCUGGCAAUUUGGC ..(..(((..(((..(((.......((((....))..(((((.((.....))))))))).((.((((((.(((....))).)))))).)).)))..))).)))..). ( -37.20) >consensus UUUUGAAUGAGUCUCCACGGAAUAAGCGCGUUCGCAUCGCUGACUAUCUGAGCAGCGGCAGAGCUCCAACUGGAGCUCCAAUUGGAGGUCAGUGGUGG________C ....(((((.((.((....))....)).))))).(((((((((((.(((((((....)).(((((((....)))))))...)))))))))))))))).......... (-35.50 = -36.25 + 0.75)



| Location | 6,345,302 – 6,345,401 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 93.37 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -30.51 |
| Energy contribution | -30.70 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.706475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6345302 99 - 22224390 G--------CCACCACUGACCUCCAAUUGGAGCUCCGGUUGGAGCUCUGCCGCUGCUCAGAUAGUCAGCGAUGCGAACGCGCUUAUUCCGUGGAGACUCAUUCAAAA .--------...((((.((.((((((((((....)))))))))).)).(((((((((.....)).)))))..((....)))).......)))).............. ( -32.60) >DroSec_CAF1 29684 99 - 1 G--------CCACCACUGACCUCCAAUUGGAGCUCCACUUGGAGCUCUGCCGCUGCUCAGAUAGUCAGCGAUGCGGACGCGCUUAUUCCGUGGAGACUCAUUCAAAA .--------.......(((.((((....((((((((....))))))))..(((((((.....)).)))))..(((((.........)))))))))..)))....... ( -32.90) >DroEre_CAF1 27572 99 - 1 G--------CCACCACUGACCUGCAAUUGGAGCUCCAAUUGGAGCUCUGCCGCUGCUCAGAUAGUCAGCGAUGCGAACGCGCUUAUUCCGUGGAGACUCAUUCAAAA .--------...((((.((...((....((((((((....))))))))..(((((((.....)).)))))..((....))))....)).)))).............. ( -32.30) >DroYak_CAF1 26305 107 - 1 GCCAAAUUGCCAGCACUGACCUCCAAUUGGAGCUCCAGUUGGAGCUCUGCCGCUGCUCAGAUAGUCAGCGAUGCGAACGCGCUUAUUCCGUGGAGACUCAUUCAAAA ................(((.((((((((((....))))))))))(((..((((((((.....)).)))))..(((....))).......)..)))......)))... ( -32.80) >consensus G________CCACCACUGACCUCCAAUUGGAGCUCCAGUUGGAGCUCUGCCGCUGCUCAGAUAGUCAGCGAUGCGAACGCGCUUAUUCCGUGGAGACUCAUUCAAAA ................(((.((((((((((....))))))))))(((..((((((((.....)).)))))..(((....))).......)..)))......)))... (-30.51 = -30.70 + 0.19)



| Location | 6,345,341 – 6,345,440 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -32.23 |
| Consensus MFE | -24.56 |
| Energy contribution | -24.25 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6345341 99 + 22224390 CUGACUAUCUGAGCAGCGGCAGAGCUCCAACCGGAGCUCCAAUUGGAGGUCAGUGGUGG--------CAAUUGGAUAAUGUUUACUAUAUUGG-CGUAGCUAAAAAAU ((((((.((..(((....)).(((((((....)))))))...)..))))))))...(((--------(...((..((((((.....)))))).-))..))))...... ( -29.00) >DroSec_CAF1 29723 98 + 1 CUGACUAUCUGAGCAGCGGCAGAGCUCCAAGUGGAGCUCCAAUUGGAGGUCAGUGGUGG--------CAAUUGGAUAAUGU-UGCUAUAUUGG-CUUAGCUAAAAAAU ((((((.((..(((....)).(((((((....)))))))...)..))))))))..((((--------((((........))-)))))).((((-(...)))))..... ( -34.60) >DroEre_CAF1 27611 98 + 1 CUGACUAUCUGAGCAGCGGCAGAGCUCCAAUUGGAGCUCCAAUUGCAGGUCAGUGGUGG--------CAAUUGGAUAAUGU-AACUAUAUUGCAAGUAGCCAAA-AAU ...((((.((((.(.((((..(((((((....)))))))...)))).).))))))))((--------(.(((.(.((((((-....))))))).))).)))...-... ( -32.20) >DroYak_CAF1 26344 105 + 1 CUGACUAUCUGAGCAGCGGCAGAGCUCCAACUGGAGCUCCAAUUGGAGGUCAGUGCUGGCAAUUUGGCAAUUGGAUAAUGU-UACCGCAUUGG-AGUAGCUAAA-AAU ((((((.((..(((....)).(((((((....)))))))...)..)))))))).........((((((.(((...((((((-....)))))).-))).))))))-... ( -33.10) >consensus CUGACUAUCUGAGCAGCGGCAGAGCUCCAACUGGAGCUCCAAUUGGAGGUCAGUGGUGG________CAAUUGGAUAAUGU_UACUAUAUUGG_AGUAGCUAAA_AAU ((((((.(((((((....)).(((((((....)))))))...))))))))))).................((((.((((((.....)))))).......))))..... (-24.56 = -24.25 + -0.31)



| Location | 6,345,341 – 6,345,440 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -26.52 |
| Consensus MFE | -20.86 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6345341 99 - 22224390 AUUUUUUAGCUACG-CCAAUAUAGUAAACAUUAUCCAAUUG--------CCACCACUGACCUCCAAUUGGAGCUCCGGUUGGAGCUCUGCCGCUGCUCAGAUAGUCAG ........((((..-......))))................--------......(((((((((((((((....)))))))))).((((........))))..))))) ( -22.90) >DroSec_CAF1 29723 98 - 1 AUUUUUUAGCUAAG-CCAAUAUAGCA-ACAUUAUCCAAUUG--------CCACCACUGACCUCCAAUUGGAGCUCCACUUGGAGCUCUGCCGCUGCUCAGAUAGUCAG ........((((..-......)))).-..............--------......(((((........((((((((....))))))))((....)).......))))) ( -23.30) >DroEre_CAF1 27611 98 - 1 AUU-UUUGGCUACUUGCAAUAUAGUU-ACAUUAUCCAAUUG--------CCACCACUGACCUGCAAUUGGAGCUCCAAUUGGAGCUCUGCCGCUGCUCAGAUAGUCAG ...-..((((((((.(((........-......((((((((--------(............)))))))))(((((....)))))........)))..)).)))))). ( -29.00) >DroYak_CAF1 26344 105 - 1 AUU-UUUAGCUACU-CCAAUGCGGUA-ACAUUAUCCAAUUGCCAAAUUGCCAGCACUGACCUCCAAUUGGAGCUCCAGUUGGAGCUCUGCCGCUGCUCAGAUAGUCAG ...-....((((((-.....((((((-........(((((....)))))........((.((((((((((....)))))))))).)))))))).....)).))))... ( -30.90) >consensus AUU_UUUAGCUACG_CCAAUAUAGUA_ACAUUAUCCAAUUG________CCACCACUGACCUCCAAUUGGAGCUCCAGUUGGAGCUCUGCCGCUGCUCAGAUAGUCAG ........((((.........))))..............................(((((((((((((((....)))))))))).((((........))))..))))) (-20.86 = -20.93 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:33:03 2006