
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,327,592 – 6,327,693 |
| Length | 101 |
| Max. P | 0.981170 |

| Location | 6,327,592 – 6,327,693 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.87 |
| Mean single sequence MFE | -24.33 |
| Consensus MFE | -12.78 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
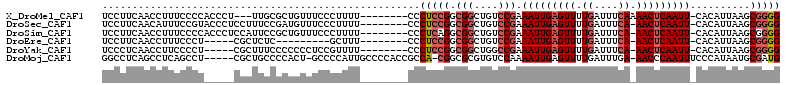
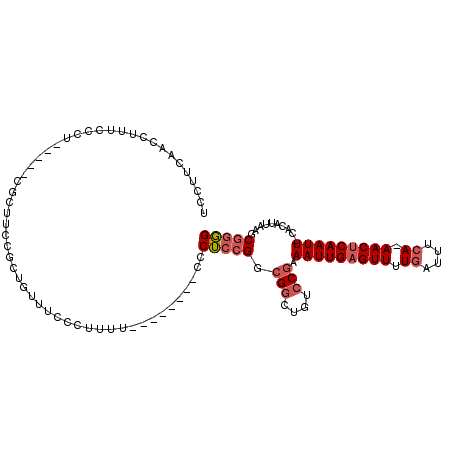
>X_DroMel_CAF1 6327592 101 + 22224390 UCCUUCAACCUUUCCCCACCCU---UUGCGCUGUUUCCCUUUU--------CCCUCCGGCGGCUGUCCGAAAUUGAGUUUUGAUUUCAAAACUCAAUU-CACAUUAAGCGGGG .............((((.....---...(((((..........--------.....)))))(((......((((((((((((....))))))))))))-.......))))))) ( -28.88) >DroSec_CAF1 12304 103 + 1 UCCUUCAACAUUUCCGUACCCUCCUUUCCGAUGUUUCCCUUUU--------CCCUCCGGCGGCUGUCCGAAAUUGAGUUUUGAUUUCA-AACUCAAUU-CACAUUAAGCGGGG ..................((((.(((...(((((.........--------.....((((....).))).((((((((((.(....))-)))))))))-.)))))))).)))) ( -19.30) >DroSim_CAF1 5705 103 + 1 UCCUUCAACCUUUCCCCACCCUCCAUUCCGCUGUUUCCCUUUU--------CCCUCAGGCGGCUGUCCGAAAUUGAGUUUUGAUUUCA-AACUCAAUU-CACAUUAAGCGGGG .............((((...........((((...........--------......))))(((......((((((((((.(....))-)))))))))-.......))))))) ( -22.55) >DroEre_CAF1 9873 89 + 1 UCCUUCAACCUUUCCCU-----CGCUCUC---------GCUUU--------CCCUCCGGCGGCUGUCCGAAAUUGAGUUUUGAUUUCA-AACUCAAUU-CACAUUAAGCGGGG ..............(((-----((((.((---------(((..--------......)))))......(((.((((((((.(....))-)))))))))-)......))))))) ( -23.30) >DroYak_CAF1 8366 98 + 1 UCCCUCAACCUUCCCCU-----CGCUUUCCCCCCCUCCGUUUU--------CCCUCCGGCGGCUGGCCGAAAUUGAGUUUUGAUUUCA-AACUCAAUU-CACAUUAAGCGGGG ..............(((-----(((((..((..((.(((....--------.....))).))..))..(((.((((((((.(....))-)))))))))-).....)))))))) ( -26.50) >DroMoj_CAF1 15466 105 + 1 GGCCUCAGCCUCAGCCU-----CGCUGCCCCACU-GCCCCAUUGCCCCACCGCCA-CGGCGCGUGUCCAAAAUUGAGUUUUGAUUUGA-AACCCAAUUUCCCAUAAUGCGAUG (((........((((..-----.)))).......-))).((((((..((((((..-..))).)))....((((((.(((((.....))-))).))))))........)))))) ( -25.46) >consensus UCCUUCAACCUUUCCCU_____CGCUUCCGCUGUUUCCCUUUU________CCCUCCGGCGGCUGUCCGAAAUUGAGUUUUGAUUUCA_AACUCAAUU_CACAUUAAGCGGGG .....................................................(((((.(((....))).(((((((((.((....)).)))))))))..........))))) (-12.78 = -13.50 + 0.72)

| Location | 6,327,592 – 6,327,693 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.87 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -13.52 |
| Energy contribution | -13.97 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
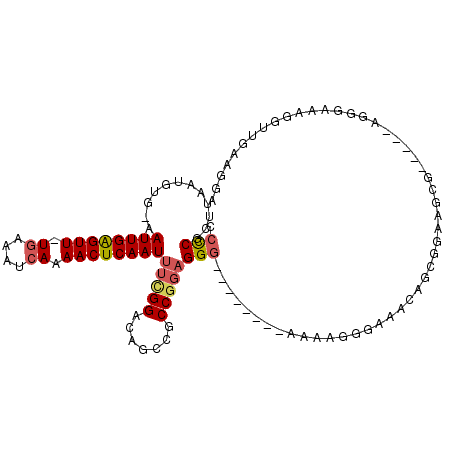
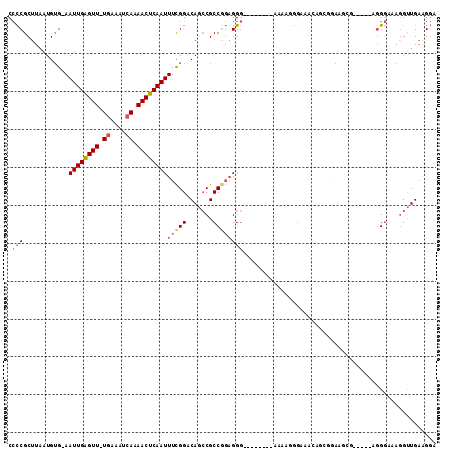
>X_DroMel_CAF1 6327592 101 - 22224390 CCCCGCUUAAUGUG-AAUUGAGUUUUGAAAUCAAAACUCAAUUUCGGACAGCCGCCGGAGGG--------AAAAGGGAAACAGCGCAA---AGGGUGGGGAAAGGUUGAAGGA ((((((((..((((-..((((((((((....))))))))))((((((.(....)))))))..--------.....(....)..)))).---.))))))))............. ( -35.10) >DroSec_CAF1 12304 103 - 1 CCCCGCUUAAUGUG-AAUUGAGUU-UGAAAUCAAAACUCAAUUUCGGACAGCCGCCGGAGGG--------AAAAGGGAAACAUCGGAAAGGAGGGUACGGAAAUGUUGAAGGA ..((..(((((((.-..(((((((-(.......))))))))(((((.((..((.((((....--------.....(....).))))...))...)).)))))))))))).)). ( -24.70) >DroSim_CAF1 5705 103 - 1 CCCCGCUUAAUGUG-AAUUGAGUU-UGAAAUCAAAACUCAAUUUCGGACAGCCGCCUGAGGG--------AAAAGGGAAACAGCGGAAUGGAGGGUGGGGAAAGGUUGAAGGA ((((((((..((((-(((((((((-(.......)))))))))))...))).((((((.....--------...))(....).))))......))))))))............. ( -32.20) >DroEre_CAF1 9873 89 - 1 CCCCGCUUAAUGUG-AAUUGAGUU-UGAAAUCAAAACUCAAUUUCGGACAGCCGCCGGAGGG--------AAAGC---------GAGAGCG-----AGGGAAAGGUUGAAGGA ((((((((..((((-(((((((((-(.......)))))))))))...)))((..((....))--------...))---------..)))))-----.)))............. ( -26.70) >DroYak_CAF1 8366 98 - 1 CCCCGCUUAAUGUG-AAUUGAGUU-UGAAAUCAAAACUCAAUUUCGGCCAGCCGCCGGAGGG--------AAAACGGAGGGGGGGAAAGCG-----AGGGGAAGGUUGAGGGA ((((((((.....(-(((((((((-(.......)))))))))))...((..((.(((.....--------....))).))..))..)))))-----.)))............. ( -32.10) >DroMoj_CAF1 15466 105 - 1 CAUCGCAUUAUGGGAAAUUGGGUU-UCAAAUCAAAACUCAAUUUUGGACACGCGCCG-UGGCGGUGGGGCAAUGGGGC-AGUGGGGCAGCG-----AGGCUGAGGCUGAGGCC ..((((.......(((((((((((-(.......))))))))))))...(((((.(((-(.((......)).)))).))-.))).....)))-----)((((........)))) ( -34.40) >consensus CCCCGCUUAAUGUG_AAUUGAGUU_UGAAAUCAAAACUCAAUUUCGGACAGCCGCCGGAGGG________AAAAGGGAAACAGCGGAAGCG_____AGGGAAAGGUUGAAGGA .(((............((((((((.((....)).))))))))(((((.......))))))))................................................... (-13.52 = -13.97 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:52 2006