
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,289,298 – 6,289,395 |
| Length | 97 |
| Max. P | 0.708315 |

| Location | 6,289,298 – 6,289,395 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -16.62 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6289298 97 + 22224390 AAAUUGAAAUGGGAACAGCGUUUACCGGCGAAAAUGGAAAUCGCCGCAAGGUGACUUGGAUUUUGGGGU---GGUUGCUCCAUGCCG--------------------AAAAGGAAUUUUG ....((((.((....))...))))(((((((.........))))).((((....))))..((((((.((---((.....)))).)))--------------------))).))....... ( -29.60) >DroSec_CAF1 26436 95 + 1 AAAUUGAAAUGGUAGCAGCGUUUACCGCCGA-AAUGGAAAUCGCCGCAAGGUGACUUGGAUUUUG-GGU---GGUUGCUCCACGUCG--------------------AAAAGGAAUUUGG ...((((..(((.((((......((((((((-(((..((.(((((....))))).))..))))).-)))---))))))))))..)))--------------------)............ ( -32.00) >DroSim_CAF1 24384 95 + 1 AAAUUGAAAUGGUAGCAGCGUUUACCGGCGA-AAUGGAAAUCGCCGCAAGGUGACUUGGAUUUUG-GGU---GGUUGCUCCACGUCG--------------------AAAAGGAAUUUGG ...((((..(((.((((((...(((((((((-........))))).((((....)))).......-)))---))))))))))..)))--------------------)............ ( -33.60) >DroYak_CAF1 23923 119 + 1 AAAUUGAAAUGGGAACAGCGUUUACCAGCGA-AAUGAAAAUCGCCGCAAGGUGACUCGGAGUUUGGCGUCUGGGCUGCUGGACGGGGCAGAUGGCUGGGUUUAUUGCGAAAGGAAUUUGG ...(((.((((((..((((((((.((.((.(-(((.....(((((....)))))......)))).))(((..(....)..)))..)).)))).))))..)))))).)))........... ( -37.00) >consensus AAAUUGAAAUGGGAACAGCGUUUACCGGCGA_AAUGGAAAUCGCCGCAAGGUGACUUGGAUUUUG_GGU___GGUUGCUCCACGUCG____________________AAAAGGAAUUUGG .........(((.((((((.....(((.......)))...(((((....)))))...................)))))))))...................................... (-16.62 = -17.50 + 0.88)

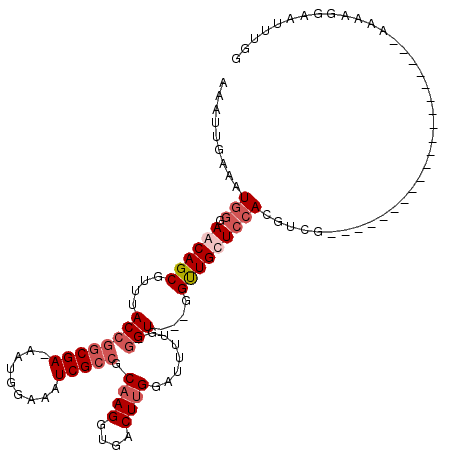
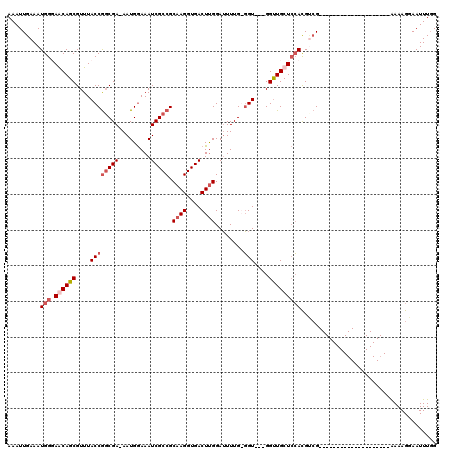
| Location | 6,289,298 – 6,289,395 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -14.69 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6289298 97 - 22224390 CAAAAUUCCUUUU--------------------CGGCAUGGAGCAACC---ACCCCAAAAUCCAAGUCACCUUGCGGCGAUUUCCAUUUUCGCCGGUAAACGCUGUUCCCAUUUCAAUUU .............--------------------.((.((((((((...---...........((((....))))((((((.........))))))........)))).)))).))..... ( -19.20) >DroSec_CAF1 26436 95 - 1 CCAAAUUCCUUUU--------------------CGACGUGGAGCAACC---ACC-CAAAAUCCAAGUCACCUUGCGGCGAUUUCCAUU-UCGGCGGUAAACGCUGCUACCAUUUCAAUUU .............--------------------.((.(((((((((((---.((-..((((..(((((.((....)).)))))..)))-).)).)))......)))).)))).))..... ( -21.40) >DroSim_CAF1 24384 95 - 1 CCAAAUUCCUUUU--------------------CGACGUGGAGCAACC---ACC-CAAAAUCCAAGUCACCUUGCGGCGAUUUCCAUU-UCGCCGGUAAACGCUGCUACCAUUUCAAUUU .............--------------------.((.((((((((...---...-.......((((....))))((((((........-))))))........)))).)))).))..... ( -23.60) >DroYak_CAF1 23923 119 - 1 CCAAAUUCCUUUCGCAAUAAACCCAGCCAUCUGCCCCGUCCAGCAGCCCAGACGCCAAACUCCGAGUCACCUUGCGGCGAUUUUCAUU-UCGCUGGUAAACGCUGUUCCCAUUUCAAUUU .......................((((....((((.......((((....(((............)))...))))(((((........-)))))))))...))))............... ( -19.10) >consensus CCAAAUUCCUUUU____________________CGACGUGGAGCAACC___ACC_CAAAAUCCAAGUCACCUUGCGGCGAUUUCCAUU_UCGCCGGUAAACGCUGCUACCAUUUCAAUUU ..................................((.((((((((.................((((....))))((((((.........))))))........)))).)))).))..... (-14.69 = -15.00 + 0.31)

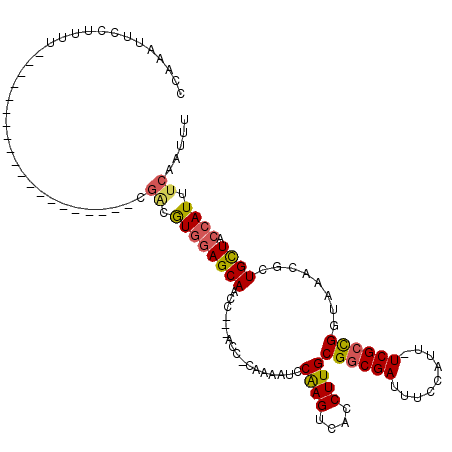
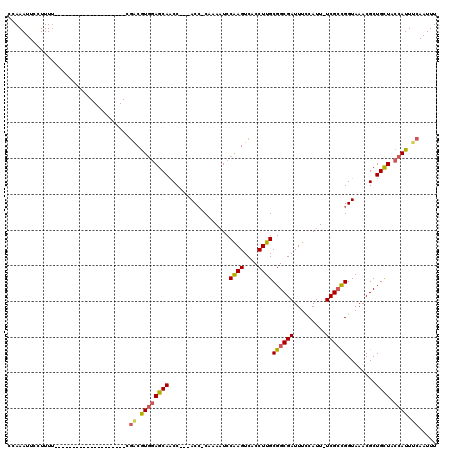
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:42 2006