
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,286,519 – 6,286,778 |
| Length | 259 |
| Max. P | 0.967438 |

| Location | 6,286,519 – 6,286,622 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.09 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -20.17 |
| Energy contribution | -20.40 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6286519 103 + 22224390 UACAAACGUAACGCAACGAAAAGUAACCAAGCAACGGGGCGUAUACGCAAUGCGAAUGCUUGACAUUACGUAUACGUAAGUAGCUGGCAAUGCAAG--AGGCAAG ......(((......)))........(((.((.((...(((((((((.(((((((....))).)))).)))))))))..)).)))))...(((...--..))).. ( -27.10) >DroSec_CAF1 23279 91 + 1 UACAAACGUAUCGAAACGAAAAGUAACCAAGCAACGGUGCGUAUACGCAAUGCGAAUGCUUGAUAUUACGUAUACGUAAGUAGCUGGC--------------AUG ......(((......)))........(((.((.((..((((((((((.(((((((....))).)))).)))))))))).)).))))).--------------... ( -23.80) >DroYak_CAF1 20399 104 + 1 UACAAAUAUAGUGCAACAGAAAGUAACCUAGCAACGGGGCGUAUACGCAAUGCAAAUGCUUGAUAUUACGCAUACGUAAGAAGCUGGCA-UGCGAGUAUGGCAAG ...........(((............(((......))).(((((.((((.(((....((((....(((((....))))).))))..)))-)))).)))))))).. ( -25.70) >consensus UACAAACGUAACGCAACGAAAAGUAACCAAGCAACGGGGCGUAUACGCAAUGCGAAUGCUUGAUAUUACGUAUACGUAAGUAGCUGGCA_UGC_AG___GGCAAG ..........................((.(((.((...(((((((((.(((((((....))).)))).)))))))))..)).))))).................. (-20.17 = -20.40 + 0.23)


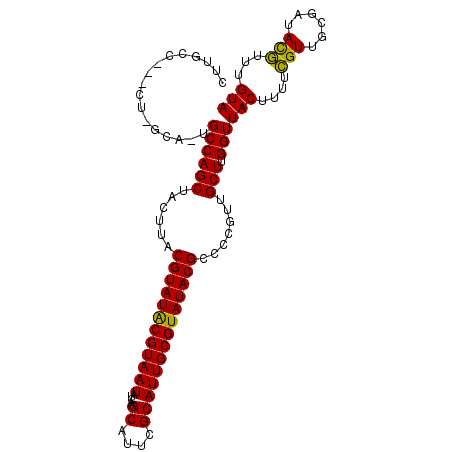
| Location | 6,286,519 – 6,286,622 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.09 |
| Mean single sequence MFE | -24.89 |
| Consensus MFE | -21.64 |
| Energy contribution | -20.75 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6286519 103 - 22224390 CUUGCCU--CUUGCAUUGCCAGCUACUUACGUAUACGUAAUGUCAAGCAUUCGCAUUGCGUAUACGCCCCGUUGCUUGGUUACUUUUCGUUGCGUUACGUUUGUA ..(((..--...)))..((((((.((...((((((((((((((.........))))))))))))))....)).)).))))(((....(((......)))...))) ( -27.40) >DroSec_CAF1 23279 91 - 1 CAU--------------GCCAGCUACUUACGUAUACGUAAUAUCAAGCAUUCGCAUUGCGUAUACGCACCGUUGCUUGGUUACUUUUCGUUUCGAUACGUUUGUA ...--------------......(((..(((((((((....((((((((..((...((((....)))).)).)))))))).......)))....))))))..))) ( -23.30) >DroYak_CAF1 20399 104 - 1 CUUGCCAUACUCGCA-UGCCAGCUUCUUACGUAUGCGUAAUAUCAAGCAUUUGCAUUGCGUAUACGCCCCGUUGCUAGGUUACUUUCUGUUGCACUAUAUUUGUA ..(((.(((......-.((((((......((((((((((((.....((....)))))))))))))).......))).))).......))).)))........... ( -23.96) >consensus CUUGCC___CU_GCA_UGCCAGCUACUUACGUAUACGUAAUAUCAAGCAUUCGCAUUGCGUAUACGCCCCGUUGCUUGGUUACUUUUCGUUGCGAUACGUUUGUA .................((((((......((((((((((((.....((....)))))))))))))).......))).)))(((....(((......)))...))) (-21.64 = -20.75 + -0.88)


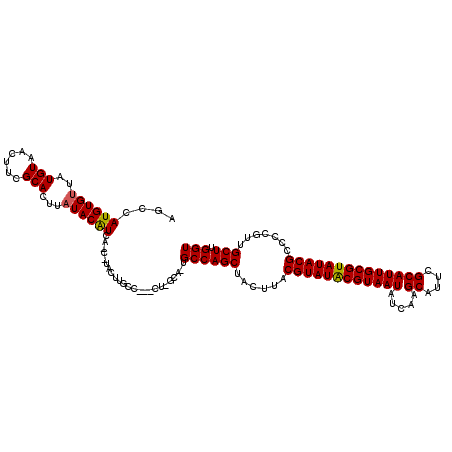
| Location | 6,286,544 – 6,286,662 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -21.63 |
| Energy contribution | -21.63 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6286544 118 + 22224390 ACCAAGCAACGGGGCGUAUACGCAAUGCGAAUGCUUGACAUUACGUAUACGUAAGUAGCUGGCAAUGCAAG--AGGCAAGUACGAUGACGUAUAAGUGCCAAGUUACAUAACACAUGGCU ....(((......(((((((((.(((((((....))).)))).)))))))))..((((((((((.(((...--..))).(((((....)))))...)))).))))))..........))) ( -37.40) >DroSec_CAF1 23304 106 + 1 ACCAAGCAACGGUGCGUAUACGCAAUGCGAAUGCUUGAUAUUACGUAUACGUAAGUAGCUGGC--------------AUGUAUGUUGAUGUAUAAGUGCGAAGUUACAUAACACAUGGCU ....(((.....((((((((((.(((((((....))).)))).)))))))))).((((((.((--------------((.((((......)))).))))..))))))..........))) ( -30.10) >DroYak_CAF1 20424 116 + 1 ACCUAGCAACGGGGCGUAUACGCAAUGCAAAUGCUUGAUAUUACGCAUACGUAAGAAGCUGGCA-UGCGAGUAUGGCAAGUA---UGAUGUAUAAGUGCAAAGCUACAUAGCACAUGGCU ....(((.((..(.(((((.((((.(((....((((....(((((....))))).))))..)))-)))).))))).)..)).---..........((((...........))))...))) ( -31.30) >consensus ACCAAGCAACGGGGCGUAUACGCAAUGCGAAUGCUUGAUAUUACGUAUACGUAAGUAGCUGGCA_UGC_AG___GGCAAGUA_G_UGAUGUAUAAGUGCAAAGUUACAUAACACAUGGCU .((.(((.((...(((((((((.(((((((....))).)))).)))))))))..)).))))).................((((............))))...(((....)))........ (-21.63 = -21.63 + 0.01)



| Location | 6,286,544 – 6,286,662 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.91 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -23.76 |
| Energy contribution | -23.65 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6286544 118 - 22224390 AGCCAUGUGUUAUGUAACUUGGCACUUAUACGUCAUCGUACUUGCCU--CUUGCAUUGCCAGCUACUUACGUAUACGUAAUGUCAAGCAUUCGCAUUGCGUAUACGCCCCGUUGCUUGGU .((((((.((.((((((...((((....((((....))))..)))).--.)))))).))))((.((...((((((((((((((.........))))))))))))))....)).)).)))) ( -37.80) >DroSec_CAF1 23304 106 - 1 AGCCAUGUGUUAUGUAACUUCGCACUUAUACAUCAACAUACAU--------------GCCAGCUACUUACGUAUACGUAAUAUCAAGCAUUCGCAUUGCGUAUACGCACCGUUGCUUGGU .....(((((((((((............))))).))))))...--------------((((((.((...((((((((((((.....((....))))))))))))))....)).)).)))) ( -26.60) >DroYak_CAF1 20424 116 - 1 AGCCAUGUGCUAUGUAGCUUUGCACUUAUACAUCA---UACUUGCCAUACUCGCA-UGCCAGCUUCUUACGUAUGCGUAAUAUCAAGCAUUUGCAUUGCGUAUACGCCCCGUUGCUAGGU ...((((((....(((.....(((..(((.....)---))..)))..))).))))-))(((((......((((((((((((.....((....)))))))))))))).......))).)). ( -26.72) >consensus AGCCAUGUGUUAUGUAACUUCGCACUUAUACAUCA_C_UACUUGCC___CU_GCA_UGCCAGCUACUUACGUAUACGUAAUAUCAAGCAUUCGCAUUGCGUAUACGCCCCGUUGCUUGGU ....((((((..(((......)))...))))))........................((((((......((((((((((((.....((....)))))))))))))).......))).))) (-23.76 = -23.65 + -0.11)


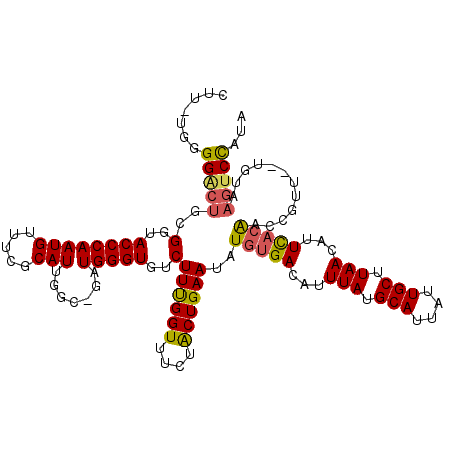
| Location | 6,286,662 – 6,286,778 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.78 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -20.15 |
| Energy contribution | -20.61 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6286662 116 - 22224390 CUUUUGGGGACUGUGGUACCCAAUGUUUCGCAUUCGC-AUUGGGUGUCUUUGGUUUAUACUGAAUAUGUGACAUUUAUGCAUUAUUGCUUAACAUUCACAACCGU---UGUAAGUCCAUA .......(((((..(..(((((((((.........))-)))))))..)((..((....))..))..(((((...(((.(((....))).)))...))))).....---....)))))... ( -31.90) >DroSec_CAF1 23410 118 - 1 CUC-UGGGGGCUGCGGUACCCAAUGUUUCGCAUUGGC-GAUGGGUGUCUUUGGUUUCUGCUGAAUAUGUGACAUUUAUGCAUUAUUGCUUAACACUCACAGCCGUUGUUGUAAGUCCAUA ...-...(((((((((((((((.....((((....))-)))))))...((..((....))..))..(((((...(((.(((....))).)))...)))))))))).......)))))... ( -32.41) >DroYak_CAF1 20540 108 - 1 CUU-UGGGGCCUGCGGUACCCAAUGU-GCGCAUUGGCUGCUGGGUGUCUUCGGUUUCUACUGAAUAUGUGACAUUUAUGCAUUAUUGCAUAACCUUUUAGAC--UU--------ACUGUA ((.-.((((((.((((...((((((.-...)))))))))).))(((((((((((....)))))).....)))))(((((((....)))))))))))..))..--..--------...... ( -32.50) >consensus CUU_UGGGGACUGCGGUACCCAAUGUUUCGCAUUGGC_GAUGGGUGUCUUUGGUUUCUACUGAAUAUGUGACAUUUAUGCAUUAUUGCUUAACAUUCACAACCGUU__UGUAAGUCCAUA .......(((((..(..((((((((.....))).......)))))..)((((((....))))))..(((((...(((.(((....))).)))...)))))............)))))... (-20.15 = -20.61 + 0.46)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:38 2006