
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,283,389 – 6,283,509 |
| Length | 120 |
| Max. P | 0.779610 |

| Location | 6,283,389 – 6,283,509 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -34.86 |
| Consensus MFE | -26.72 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
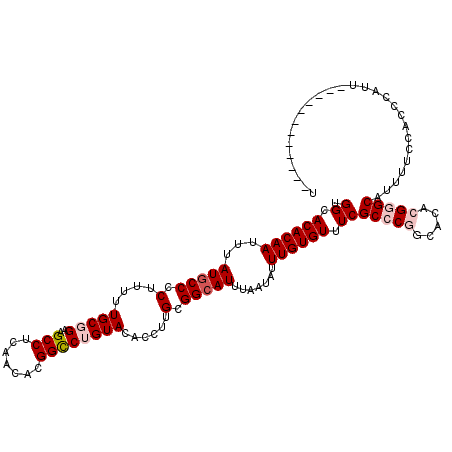
>X_DroMel_CAF1 6283389 120 + 22224390 AAAUGGUUGGAAAAAUGGGUGGAAAAUGCCCGUGUGCCGGGCGAAACACAAAUAUUAAAUCCCGCAAGGUGUACAGGCCGUGUUGAGGCUUCCGCAAAAAGGGGCAUAAAUUGUGUGCCA .......(((....(((((((.....)))))))...)))(((...((((((...(((..((((.....(((...(((((.......))))).))).....))))..))).))))))))). ( -34.70) >DroPse_CAF1 5139 98 + 1 -------------UGUGG-------CUGCUC--GUGCUGGGCGAAACACAAAUAUUAAAUGCCGCAAGGUGUACGGACCGUGUUGAGGCUCCGGCAAAAAGGGGCAUAAAUUGUGUGCCC -------------((((.-------.(((((--.....)))))...))))........(((((....))))).((((((.......)).))))........(((((((.....))))))) ( -32.80) >DroSec_CAF1 20375 120 + 1 AAAUGGUUGGAAAAAUGGGUGGAAAAUGCCCGUGUGCCGGGCGAAACACAAAUAUUAAAUGCCGCAAGGUGUACAGGCCGUGUUGAGGCUUCCGCAAAAAGGGGCAUAAAUUGUGUGCCA .......(((....(((((((.....)))))))...)))(((...((((((.......(((((.(....(((..(((((.......)))))..)))....).)))))...))))))))). ( -36.40) >DroPer_CAF1 5372 98 + 1 -------------UGUGG-------CUGCUC--GUGCUGGGCGAAACACAAAUAUUAAAUGCCGCAAGGUGUACGGACCGUGUUGAGGCUCCGGCAAAAGGGGGCAUAAAUUGUGUGCCC -------------.....-------...((.--.((((((((..(((((.........(((((....))))).......)))))...)).))))))..)).(((((((.....))))))) ( -34.19) >DroYak_CAF1 17476 108 + 1 A------------AAUGGGUGGAAAAUGCCCGUGUGCCGGGCGAAACACAAAUAUUAAAUGCCGCAAGGUGUACAGGCCGUGUUGAGGCUUCCGCAAAAAGGGGCAUAAAUUGUGUGCCA .------------.....((((((..((((((.....))))))...............(((((....)))))....(((.......))))))))).......((((((.....)))))). ( -36.20) >consensus A____________AAUGGGUGGAAAAUGCCCGUGUGCCGGGCGAAACACAAAUAUUAAAUGCCGCAAGGUGUACAGGCCGUGUUGAGGCUUCCGCAAAAAGGGGCAUAAAUUGUGUGCCA ................((........((((((.....))))))..((((((.......(((((.(....(((..(((((.......)))))..)))....).)))))...)))))).)). (-26.72 = -27.00 + 0.28)



| Location | 6,283,389 – 6,283,509 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -21.08 |
| Energy contribution | -22.24 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6283389 120 - 22224390 UGGCACACAAUUUAUGCCCCUUUUUGCGGAAGCCUCAACACGGCCUGUACACCUUGCGGGAUUUAAUAUUUGUGUUUCGCCCGGCACACGGGCAUUUUCCACCCAUUUUUCCAACCAUUU (((.((((((.(((..((((....(((((..(((.......))))))))......).)))...)))...))))))...(((((.....))))).....................)))... ( -29.70) >DroPse_CAF1 5139 98 - 1 GGGCACACAAUUUAUGCCCCUUUUUGCCGGAGCCUCAACACGGUCCGUACACCUUGCGGCAUUUAAUAUUUGUGUUUCGCCCAGCAC--GAGCAG-------CCACA------------- ((((((((((...(((((.(....((.((((.((.......))))))..))....).))))).......))))))...)))).((..--..))..-------.....------------- ( -26.20) >DroSec_CAF1 20375 120 - 1 UGGCACACAAUUUAUGCCCCUUUUUGCGGAAGCCUCAACACGGCCUGUACACCUUGCGGCAUUUAAUAUUUGUGUUUCGCCCGGCACACGGGCAUUUUCCACCCAUUUUUCCAACCAUUU (((.((((((...(((((.(....(((((..(((.......))))))))......).))))).......))))))...(((((.....))))).....................)))... ( -30.20) >DroPer_CAF1 5372 98 - 1 GGGCACACAAUUUAUGCCCCCUUUUGCCGGAGCCUCAACACGGUCCGUACACCUUGCGGCAUUUAAUAUUUGUGUUUCGCCCAGCAC--GAGCAG-------CCACA------------- ((((((((((...((((((........((((.((.......))))))........).))))).......))))))...)))).((..--..))..-------.....------------- ( -25.89) >DroYak_CAF1 17476 108 - 1 UGGCACACAAUUUAUGCCCCUUUUUGCGGAAGCCUCAACACGGCCUGUACACCUUGCGGCAUUUAAUAUUUGUGUUUCGCCCGGCACACGGGCAUUUUCCACCCAUU------------U .((.((((((...(((((.(....(((((..(((.......))))))))......).))))).......)))))).))(((((.....)))))..............------------. ( -30.00) >consensus UGGCACACAAUUUAUGCCCCUUUUUGCGGAAGCCUCAACACGGCCUGUACACCUUGCGGCAUUUAAUAUUUGUGUUUCGCCCGGCACACGGGCAUUUUCCACCCAUU____________U .((.((((((...(((((.(....(((((..(((.......))))))))......).))))).......)))))).))(((((.....)))))........................... (-21.08 = -22.24 + 1.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:30 2006