
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,283,134 – 6,283,250 |
| Length | 116 |
| Max. P | 0.999596 |

| Location | 6,283,134 – 6,283,250 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.95 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -30.57 |
| Energy contribution | -31.37 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6283134 116 + 22224390 AAUUGCGGCCAGG----CAGGUCCUUGGCCCCCAGGACAUGCGGUCAAUCAUAAAUUGCCUUAGCGAAAGCCUUCGCCAAAGCGUAAACAAGUCGACGCAGUCCUGACAUUAUUAUAUUU ......(((((((----......)))))))..((((((.((((.((..........(((.((.(((((....))))).)).)))..........)))))))))))).............. ( -35.95) >DroPse_CAF1 4415 117 + 1 AAUUGCGGCCCGG---CCAGGUCCUUGGCCCCCAGGACAUGCGGUCAAUCAUAAAUUGCCUUAGCGAAAGCCUUCGCCAAAGCGUAAACAAGUCGACGCAGACCUGACAUUAUUAUAUUU ..(((((...(((---(...(((((.(....).)))))((((((.((((.....))))))...(((((....)))))....))))......)))).)))))................... ( -30.30) >DroSec_CAF1 20148 116 + 1 AAUUGCGGCCAGG----CAGGUCCUUGGCCCCCAGGACAUGCGGUCAAUCAUAAAUUGCCUUCGCGAAAGCCUUCGCCAAAGCGUAAACAAGUCGACGCAGUCCUGACAUUAUUAUAUUU ......(((((((----......)))))))..((((((.((((.((..........(((.((.(((((....))))).)).)))..........)))))))))))).............. ( -35.95) >DroPer_CAF1 4664 117 + 1 AAUUGCGGCCCGG---CCAGGUCCUUGGCCCCCAGGACAUGCGGUCAAUCAUAAAUUGCCUUAGCGAAAGCCUUCGCCAAAGCGUAAACAAGUCGACGCAGACCUGACAUUAUUAUAUUU ..(((((...(((---(...(((((.(....).)))))((((((.((((.....))))))...(((((....)))))....))))......)))).)))))................... ( -30.30) >DroYak_CAF1 17229 120 + 1 AAUUGCGGCCAGGCGGGCAGGUCCUUGGCCCCCAGGACAUGCGGUCAAUCAUAAAUUGCCUUAGCGAAAGCCUUCGCCAAAGCGUAAACAAGUCGACGCAGUCCUGACAUUAUUAUAUUU ......(((((((.((.....)))))))))..((((((.((((.((..........(((.((.(((((....))))).)).)))..........)))))))))))).............. ( -36.35) >consensus AAUUGCGGCCAGG____CAGGUCCUUGGCCCCCAGGACAUGCGGUCAAUCAUAAAUUGCCUUAGCGAAAGCCUUCGCCAAAGCGUAAACAAGUCGACGCAGUCCUGACAUUAUUAUAUUU ......(((((((..........)))))))..((((((.((((.((..........(((.((.(((((....))))).)).)))..........)))))))))))).............. (-30.57 = -31.37 + 0.80)

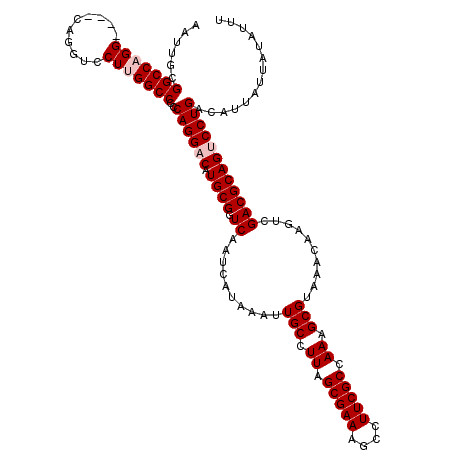

| Location | 6,283,134 – 6,283,250 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.95 |
| Mean single sequence MFE | -43.52 |
| Consensus MFE | -39.88 |
| Energy contribution | -40.88 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6283134 116 - 22224390 AAAUAUAAUAAUGUCAGGACUGCGUCGACUUGUUUACGCUUUGGCGAAGGCUUUCGCUAAGGCAAUUUAUGAUUGACCGCAUGUCCUGGGGGCCAAGGACCUG----CCUGGCCGCAAUU ..............((((((((((((((.(..(....(((((((((((....))))))))))).....)..))))).)))).))))))(.(((((.((.....----))))))).).... ( -46.30) >DroPse_CAF1 4415 117 - 1 AAAUAUAAUAAUGUCAGGUCUGCGUCGACUUGUUUACGCUUUGGCGAAGGCUUUCGCUAAGGCAAUUUAUGAUUGACCGCAUGUCCUGGGGGCCAAGGACCUGG---CCGGGCCGCAAUU ..............((((.(((((((((.(..(....(((((((((((....))))))))))).....)..))))).)))).).))))(.((((..((......---)).)))).).... ( -40.50) >DroSec_CAF1 20148 116 - 1 AAAUAUAAUAAUGUCAGGACUGCGUCGACUUGUUUACGCUUUGGCGAAGGCUUUCGCGAAGGCAAUUUAUGAUUGACCGCAUGUCCUGGGGGCCAAGGACCUG----CCUGGCCGCAAUU ..............((((((((((((((.(..(....(((((.(((((....))))).))))).....)..))))).)))).))))))(.(((((.((.....----))))))).).... ( -44.20) >DroPer_CAF1 4664 117 - 1 AAAUAUAAUAAUGUCAGGUCUGCGUCGACUUGUUUACGCUUUGGCGAAGGCUUUCGCUAAGGCAAUUUAUGAUUGACCGCAUGUCCUGGGGGCCAAGGACCUGG---CCGGGCCGCAAUU ..............((((.(((((((((.(..(....(((((((((((....))))))))))).....)..))))).)))).).))))(.((((..((......---)).)))).).... ( -40.50) >DroYak_CAF1 17229 120 - 1 AAAUAUAAUAAUGUCAGGACUGCGUCGACUUGUUUACGCUUUGGCGAAGGCUUUCGCUAAGGCAAUUUAUGAUUGACCGCAUGUCCUGGGGGCCAAGGACCUGCCCGCCUGGCCGCAAUU ..............((((((((((((((.(..(....(((((((((((....))))))))))).....)..))))).)))).))))))(.(((((.((......))...))))).).... ( -46.10) >consensus AAAUAUAAUAAUGUCAGGACUGCGUCGACUUGUUUACGCUUUGGCGAAGGCUUUCGCUAAGGCAAUUUAUGAUUGACCGCAUGUCCUGGGGGCCAAGGACCUG____CCUGGCCGCAAUU ..............((((((((((((((.(..(....(((((((((((....))))))))))).....)..))))).)))).))))))(.(((((.((.........))))))).).... (-39.88 = -40.88 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:28 2006