
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,272,744 – 6,272,889 |
| Length | 145 |
| Max. P | 0.999537 |

| Location | 6,272,744 – 6,272,856 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.70 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -26.97 |
| Energy contribution | -27.05 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6272744 112 - 22224390 UCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGGGUU-------UCUCAGGAUUUUUCCG-GCUCUUCCGGCUUUUU .......(((((((((.(((((.....)))))))..((((.((......)))))).)))))))((((((((.((((((..-------((....))...)))))-)...)))))))).... ( -34.90) >DroSec_CAF1 15029 110 - 1 UCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGGGUU-------UUUCAGGGUU-UUCCG-GCUCUUCCGGCUU-CU .......(((((((((.(((((.....)))))))..((((.((......)))))).)))))))((((((((.((((((..-------((....))..-)))))-)...)))))))).-.. ( -33.50) >DroSim_CAF1 16587 110 - 1 UCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGGGUU-------UUUCAGGGUU-UUCCG-GCUCUUCCGGCUU-CU .......(((((((((.(((((.....)))))))..((((.((......)))))).)))))))((((((((.((((((..-------((....))..-)))))-)...)))))))).-.. ( -33.50) >DroEre_CAF1 4042 112 - 1 UCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAACCGGGGUCCGGGGUU-------UUUCAGGGGUUUUCAGGGUUUUUCCGGCUU-CU ......(((((..(((.(((((.....))))))))............(((((...)))))..)))))((((.((((((..-------..((..((....))..))...)))))))))-). ( -27.70) >DroYak_CAF1 12228 118 - 1 UCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGAGUUCUUUUUUUUUCCAGGCUUUUCCG-GCUUCUCCGGCUU-CU .......(((((((((.(((((.....)))))))..((((.((......)))))).)))))))((((((((.((((((..(((.........)))...)))))-)...)))))))).-.. ( -36.70) >consensus UCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGGGUU_______UUUCAGGGUUUUUCCG_GCUCUUCCGGCUU_CU .......(((((((((.(((((.....)))))))..((((.((......)))))).)))))))((((((((.((((((....................))))).)...)))))))).... (-26.97 = -27.05 + 0.08)



| Location | 6,272,776 – 6,272,889 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.34 |
| Mean single sequence MFE | -36.84 |
| Consensus MFE | -35.06 |
| Energy contribution | -35.18 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6272776 113 - 22224390 GCG-------GCACUUGCGGUCGAAAGGCAAACUCUGAGUUCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGGGUU ...-------((....)).(((....))).(((((((..(((.((((.((((((((.(((((.....)))))))..((((.((......)))))).))))))))))..)))..))))))) ( -31.90) >DroSec_CAF1 15059 113 - 1 GCG-------GCACUUGCGGUCGAAAGACAAACUCUGGGUUCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGGGUU ...-------((....)).(((....))).(((((((((..(.((((.((((((((.(((((.....)))))))..((((.((......)))))).))))))))))..)..))))))))) ( -37.80) >DroSim_CAF1 16617 113 - 1 GCG-------GCACUUGCGGUCGAAAGGCAAACUCUGGGUUCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGGGUU ...-------((....)).(((....))).(((((((((..(.((((.((((((((.(((((.....)))))))..((((.((......)))))).))))))))))..)..))))))))) ( -37.20) >DroEre_CAF1 4074 113 - 1 GCG-------GCACUUGCGGUCGAAAGGCAAACUCGGGGUUCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAACCGGGGUCCGGGGUU ...-------((....)).(((....)))...((((((.(((.((((.((((((((.(((((.....)))))))..((((.((......)))))).)))))))))).))).))))))... ( -35.50) >DroYak_CAF1 12266 120 - 1 GCUGCAAGGAGCACUUGCGGUCGAAAGGCAAACUCGGGGUUCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGAGUU ..((((((.....))))))(((....))).(((((.(((..(.((((.((((((((.(((((.....)))))))..((((.((......)))))).))))))))))..)..))).))))) ( -41.80) >consensus GCG_______GCACUUGCGGUCGAAAGGCAAACUCUGGGUUCAGUUGGUUGAGUGAAUGGAUUAAAAGUCCAUCAGAAUGUACAUAAAAGUCAUUGACUUAACAGCCGGGGUCCGGGGUU ..........((....)).(((....))).(((((((((..(.((((.((((((((.(((((.....)))))))..((((.((......)))))).))))))))))..)..))))))))) (-35.06 = -35.18 + 0.12)

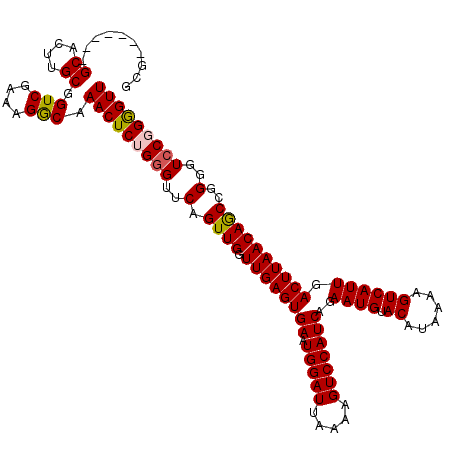
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:24 2006