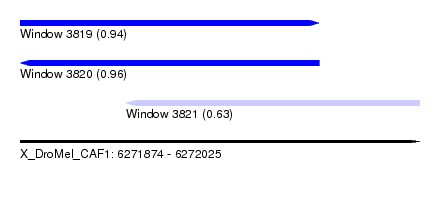
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,271,874 – 6,272,025 |
| Length | 151 |
| Max. P | 0.961812 |

| Location | 6,271,874 – 6,271,987 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.22 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6271874 113 + 22224390 GCAUUGCACAACAUUGAACAUCAUGCUCGGCUCAUUACAGAAAGCCCCGUUUCAUGGCCAACAAAAUGUUGGC-------CAGGUAAUAAUUCAAAUUGAUGACCGACAUUCGAUCGCAU ....(((.....(((((.....(((.((((.((((((..((((......)))).((((((((.....))))))-------))...............)))))))))))))))))).))). ( -28.90) >DroSec_CAF1 14204 113 + 1 GCAUUGCACAACAUUGAACAUCAUGCUCGGCUCAUUACAAAAAGCCCCGUUUCAUGGCCAACAAAAUGUUGGC-------CAGGUAAUAAUUCAAAUUGAUGACCGACAUUCGAUCGCAU ....(((.....(((((.....(((.((((.((((((...........(((...((((((((.....))))))-------))...))).........)))))))))))))))))).))). ( -26.85) >DroSim_CAF1 15752 113 + 1 GCAUUGCACAACAUUGAACAUCAUGCUCGGCUCAUUACAAAAAGCCCCGUUUCAUGGCCAACAAAAUGUUGGC-------CAGGUAAUAAUUCAAAUUGAUGACCGACAUUCGAUCGCAU ....(((.....(((((.....(((.((((.((((((...........(((...((((((((.....))))))-------))...))).........)))))))))))))))))).))). ( -26.85) >DroEre_CAF1 3215 113 + 1 ACAUUGCACAACAUUGAACAUCAUGCUCGGCUCAUUACAAAAAGCACUGUUUCAUGGCCAACAAAAUGUUGGC-------CAGGUAAUAAUUCAAAUUGAUGACCGACAUUCGAUCGCAU ....(((.....(((((.....(((.((((.((((((..........((((...((((((((.....))))))-------))...))))........)))))))))))))))))).))). ( -27.97) >DroYak_CAF1 11367 120 + 1 AUAUUGCACAACAUUGAACAUCAUGCUCGGCUCAUUACGAAAAAUCCCGUUUCAUGGCCAACAAAAUGUUGGUACAGGUACAGGUUAUAAUUCAAAUUCAUGACCGACAUUCGAUCGCAU ....(((.(((((((((.(.....).))(((.(((.(((........)))...)))))).....)))))))......((...(((((((((....))).)))))).))........))). ( -23.70) >consensus GCAUUGCACAACAUUGAACAUCAUGCUCGGCUCAUUACAAAAAGCCCCGUUUCAUGGCCAACAAAAUGUUGGC_______CAGGUAAUAAUUCAAAUUGAUGACCGACAUUCGAUCGCAU ....(((.....(((((.....(((.((((.((((((((..((((...))))..))((((((.....))))))........................)))))))))))))))))).))). (-21.66 = -21.22 + -0.44)

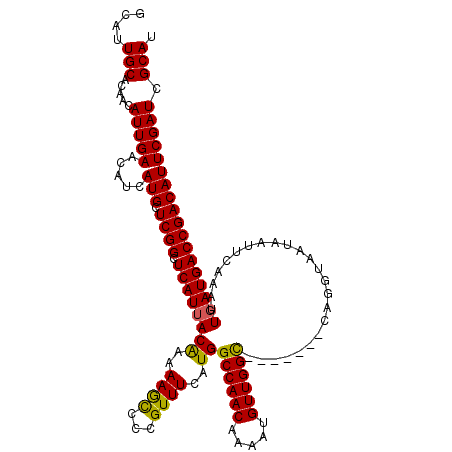

| Location | 6,271,874 – 6,271,987 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -34.16 |
| Consensus MFE | -27.58 |
| Energy contribution | -28.22 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6271874 113 - 22224390 AUGCGAUCGAAUGUCGGUCAUCAAUUUGAAUUAUUACCUG-------GCCAACAUUUUGUUGGCCAUGAAACGGGGCUUUCUGUAAUGAGCCGAGCAUGAUGUUCAAUGUUGUGCAAUGC .((((((((.....)))))..((((((((((.(((((.((-------((((((.....)))))))).)......(((((........))))).....)))))))))).)))).))).... ( -36.30) >DroSec_CAF1 14204 113 - 1 AUGCGAUCGAAUGUCGGUCAUCAAUUUGAAUUAUUACCUG-------GCCAACAUUUUGUUGGCCAUGAAACGGGGCUUUUUGUAAUGAGCCGAGCAUGAUGUUCAAUGUUGUGCAAUGC .((((((((.....)))))..((((((((((.(((((.((-------((((((.....)))))))).)......(((((........))))).....)))))))))).)))).))).... ( -36.30) >DroSim_CAF1 15752 113 - 1 AUGCGAUCGAAUGUCGGUCAUCAAUUUGAAUUAUUACCUG-------GCCAACAUUUUGUUGGCCAUGAAACGGGGCUUUUUGUAAUGAGCCGAGCAUGAUGUUCAAUGUUGUGCAAUGC .((((((((.....)))))..((((((((((.(((((.((-------((((((.....)))))))).)......(((((........))))).....)))))))))).)))).))).... ( -36.30) >DroEre_CAF1 3215 113 - 1 AUGCGAUCGAAUGUCGGUCAUCAAUUUGAAUUAUUACCUG-------GCCAACAUUUUGUUGGCCAUGAAACAGUGCUUUUUGUAAUGAGCCGAGCAUGAUGUUCAAUGUUGUGCAAUGU .((((((((.....)))))..((((((((((.(((.(.((-------((((((.....)))))))).).....((((((...((.....)).))))))))))))))).)))).))).... ( -33.20) >DroYak_CAF1 11367 120 - 1 AUGCGAUCGAAUGUCGGUCAUGAAUUUGAAUUAUAACCUGUACCUGUACCAACAUUUUGUUGGCCAUGAAACGGGAUUUUUCGUAAUGAGCCGAGCAUGAUGUUCAAUGUUGUGCAAUAU .((((((((.....))))).............(((((..(((....))).((((((.((((((((((...(((((....))))).))).))).)))).))))))....)))))))).... ( -28.70) >consensus AUGCGAUCGAAUGUCGGUCAUCAAUUUGAAUUAUUACCUG_______GCCAACAUUUUGUUGGCCAUGAAACGGGGCUUUUUGUAAUGAGCCGAGCAUGAUGUUCAAUGUUGUGCAAUGC .((((((((.....)))))..((((((((((................((((((.....))))))((((......(((((........)))))...))))..)))))).)))).))).... (-27.58 = -28.22 + 0.64)

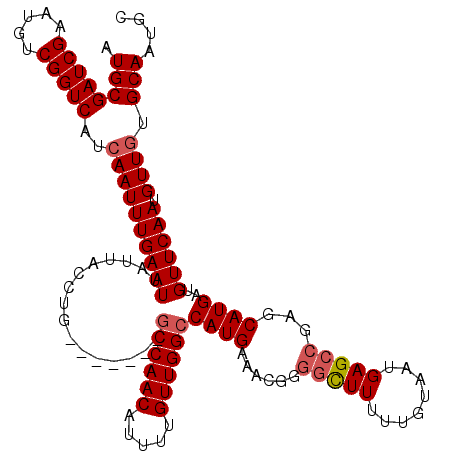

| Location | 6,271,914 – 6,272,025 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.36 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -26.90 |
| Energy contribution | -27.14 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6271914 111 - 22224390 GGACAAGUGAGCCUGAUUGGGGCUACCGCAAA--UUCGGGAUGCGAUCGAAUGUCGGUCAUCAAUUUGAAUUAUUACCUG-------GCCAACAUUUUGUUGGCCAUGAAACGGGGCUUU ........((((((((((((.((..(((....--..)))...))(((((.....))))).))))))..........(.((-------((((((.....)))))))).).....)))))). ( -36.30) >DroSec_CAF1 14244 111 - 1 GGACAAGUGAGCCUGAUUGGGGCUCCCGCAAA--UUCGGGAUGCGAUCGAAUGUCGGUCAUCAAUUUGAAUUAUUACCUG-------GCCAACAUUUUGUUGGCCAUGAAACGGGGCUUU ........((((((((((((.(((((((....--..))))).))(((((.....))))).))))))..........(.((-------((((((.....)))))))).).....)))))). ( -40.80) >DroSim_CAF1 15792 111 - 1 GGACAAGUGAGCCUGAUUGGGGCUCCCGCAAA--UUCGGGAUGCGAUCGAAUGUCGGUCAUCAAUUUGAAUUAUUACCUG-------GCCAACAUUUUGUUGGCCAUGAAACGGGGCUUU ........((((((((((((.(((((((....--..))))).))(((((.....))))).))))))..........(.((-------((((((.....)))))))).).....)))))). ( -40.80) >DroEre_CAF1 3255 111 - 1 AGACAAGUGAGCCUGAUUGUGCCUCCCGCAAG--CUCGGGAUGCGAUCGAAUGUCGGUCAUCAAUUUGAAUUAUUACCUG-------GCCAACAUUUUGUUGGCCAUGAAACAGUGCUUU ........(((((((((((.((.(((((....--..))))).))(((((.....)))))..))))...........(.((-------((((((.....)))))))).)...))).)))). ( -38.20) >DroYak_CAF1 11407 120 - 1 GGACAAGUGAGCCUGAUUGUGCCAGCCGCAAAAUUUCGGGAUGCGAUCGAAUGUCGGUCAUGAAUUUGAAUUAUAACCUGUACCUGUACCAACAUUUUGUUGGCCAUGAAACGGGAUUUU ...((((((..(((((...(((.....))).....)))))...)(((((.....)))))....)))))........(((((..(....(((((.....)))))....)..)))))..... ( -29.30) >consensus GGACAAGUGAGCCUGAUUGGGGCUCCCGCAAA__UUCGGGAUGCGAUCGAAUGUCGGUCAUCAAUUUGAAUUAUUACCUG_______GCCAACAUUUUGUUGGCCAUGAAACGGGGCUUU ...((((((..((((((((.(.....).)))....)))))...)(((((.....)))))....)))))........((((.......((((((.....)))))).......))))..... (-26.90 = -27.14 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:22 2006