| Sequence ID | X_DroMel_CAF1 |
|---|---|
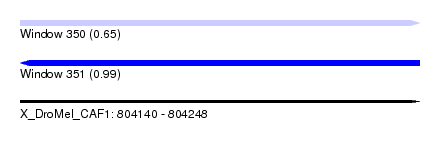
| Location | 804,140 – 804,248 |
| Length | 108 |
| Max. P | 0.987189 |

| Location | 804,140 – 804,248 |
|---|---|
| Length | 108 |
| Sequences | 6 |
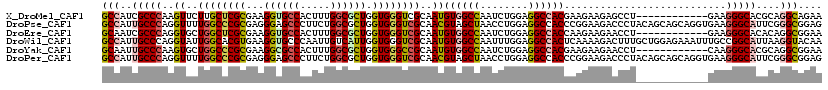
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -43.87 |
| Consensus MFE | -24.20 |
| Energy contribution | -24.95 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
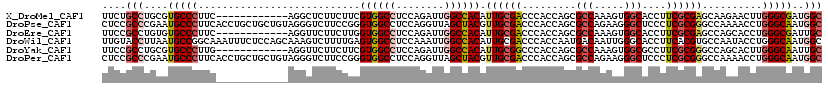
>X_DroMel_CAF1 804140 108 + 22224390 UUCUGCCUGCGUGCCCUUC------------AGGCUCUUCUUCGUGGCCUCCAGAUUGGCCACAUUGCGACCCACCAGCGCCAAAGUGGCACCUUCGCGAGCAAGAACUUGGGCGAUGGC ....(((....((((((((------------..((((......((((((........))))))...((((.......(.(((.....))).)..))))))))..)))...)))))..))) ( -40.60) >DroPse_CAF1 601 120 + 1 CUCCGCCCGAAUGCCCUUCACCUGCUGCUGUAGGGUCUUCCGGGUGGCCUCCAGGUUAGCUACGUUGCGACCCACCAGCGCCAGAAGGGCUCCCUCGCGGGCCAAAACCUGGGCAAUGGC ....(((((...(((((((....((.((((..(((((.......(((((....)))))((......)))))))..))))))..))))))).(((....)))........)))))...... ( -48.50) >DroEre_CAF1 592 108 + 1 UUCCGCCUGUGUGCCCUUC------------AGGUUCUUCUUGGUGGCCUCCAGAUUGGCCACAUUGCGACCCACCAGCGCCAAAGUGGCACCUUCGCGAGCCAGCACCUGGGCGAUUGC ...((((((.((((.....------------.(((((......((((((........))))))...((((.......(.(((.....))).)..))))))))).)))).))))))..... ( -39.90) >DroWil_CAF1 592 120 + 1 UUGUACCUUAAUGCCGGCAAAUUUCUCCAGCAAAGUCUUUUGAGUGGCCUCCAAAUUGGCCACAUUGCGACCCACCAAUGACAAUUGGGCACCUUCACGUGCCAAUACCUGGGCAAUGGC ............((((.............((((..((....))((((((........)))))).))))(.((((.((((....))))(((((......)))))......)))))..)))) ( -35.80) >DroYak_CAF1 592 108 + 1 UUCCGCCUGCGUGCCCUUG------------AGGUUCUUCUUCGUGGCCUCCAGAUUGGCCACAUUGCGGCCCACCAGCGCCAAAGUGGCGCCUUCGCGGGCCAGCACUUGGGCAAUUGC ....(((((.((((....(------------(((.....))))((((((........)))))).....(((((.(..(((((.....)))))....).))))).)))).)))))...... ( -49.90) >DroPer_CAF1 601 120 + 1 CUCCGCCCGAAUGCCCUUCACCUGCUGCUGUAGGGUCUUCCGGGUGGCCUCCAGGUUAGCUACGUUGCGACCCACCAGCGCCAGAAGGGCUCCCUCGCGGGCCAAAACCUGGGCAAUGGC ....(((((...(((((((....((.((((..(((((.......(((((....)))))((......)))))))..))))))..))))))).(((....)))........)))))...... ( -48.50) >consensus UUCCGCCUGAAUGCCCUUC____________AGGGUCUUCUGGGUGGCCUCCAGAUUGGCCACAUUGCGACCCACCAGCGCCAAAGGGGCACCUUCGCGGGCCAAAACCUGGGCAAUGGC ....(((....(((((...........................((((((........)))))).((((((.........(((.....)))....))))))..........)))))..))) (-24.20 = -24.95 + 0.75)
| Location | 804,140 – 804,248 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.57 |
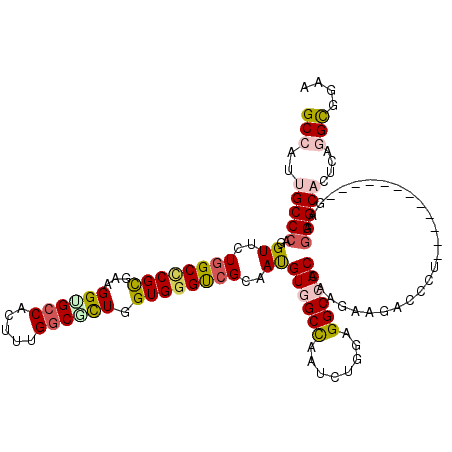
| Mean single sequence MFE | -50.70 |
| Consensus MFE | -36.16 |
| Energy contribution | -36.83 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
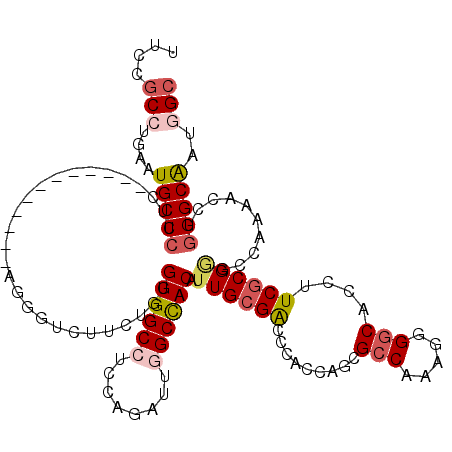
>X_DroMel_CAF1 804140 108 - 22224390 GCCAUCGCCCAAGUUCUUGCUCGCGAAGGUGCCACUUUGGCGCUGGUGGGUCGCAAUGUGGCCAAUCUGGAGGCCACGAAGAAGAGCCU------------GAAGGGCACGCAGGCAGAA (((...((((...(((..((((((((.((((((.....))))))......))))..(((((((........))))))).....))))..------------))))))).....))).... ( -46.80) >DroPse_CAF1 601 120 - 1 GCCAUUGCCCAGGUUUUGGCCCGCGAGGGAGCCCUUCUGGCGCUGGUGGGUCGCAACGUAGCUAACCUGGAGGCCACCCGGAAGACCCUACAGCAGCAGGUGAAGGGCAUUCGGGCGGAG ((((..((....))..))))((((..(((.(((((((..((((((..(((((((......))....((((.(....)))))..)))))..)))).))....))))))).)))..)))).. ( -57.30) >DroEre_CAF1 592 108 - 1 GCAAUCGCCCAGGUGCUGGCUCGCGAAGGUGCCACUUUGGCGCUGGUGGGUCGCAAUGUGGCCAAUCUGGAGGCCACCAAGAAGAACCU------------GAAGGGCACACAGGCGGAA ....(((((...((((((((((((...((((((.....)))))).))))))).....((((((........))))))............------------....)))))...))))).. ( -46.90) >DroWil_CAF1 592 120 - 1 GCCAUUGCCCAGGUAUUGGCACGUGAAGGUGCCCAAUUGUCAUUGGUGGGUCGCAAUGUGGCCAAUUUGGAGGCCACUCAAAAGACUUUGCUGGAGAAAUUUGCCGGCAUUAAGGUACAA ((((.(((....))).))))..(((..(((((((((...((.(..(..((((.....((((((........))))))......))).)..)..).))...)))..))))))....))).. ( -41.80) >DroYak_CAF1 592 108 - 1 GCAAUUGCCCAAGUGCUGGCCCGCGAAGGCGCCACUUUGGCGCUGGUGGGCCGCAAUGUGGCCAAUCUGGAGGCCACGAAGAAGAACCU------------CAAGGGCACGCAGGCGGAA ((...(((((...(((.(((((((...((((((.....)))))).)))))))))).(((((((........)))))))...........------------...))))).....)).... ( -54.10) >DroPer_CAF1 601 120 - 1 GCCAUUGCCCAGGUUUUGGCCCGCGAGGGAGCCCUUCUGGCGCUGGUGGGUCGCAACGUAGCUAACCUGGAGGCCACCCGGAAGACCCUACAGCAGCAGGUGAAGGGCAUUCGGGCGGAG ((((..((....))..))))((((..(((.(((((((..((((((..(((((((......))....((((.(....)))))..)))))..)))).))....))))))).)))..)))).. ( -57.30) >consensus GCCAUUGCCCAGGUUCUGGCCCGCGAAGGUGCCACUUUGGCGCUGGUGGGUCGCAAUGUGGCCAAUCUGGAGGCCACCAAGAAGACCCU____________GAAGGGCACUCAGGCGGAA (((..(((((..((..((((((((...((((((.....)))))).))))))))..))((((((........))))))...........................)))))....))).... (-36.16 = -36.83 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:34 2006