
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,266,488 – 6,266,630 |
| Length | 142 |
| Max. P | 0.980417 |

| Location | 6,266,488 – 6,266,590 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.64 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -20.79 |
| Energy contribution | -20.60 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6266488 102 + 22224390 UAAUGGGCGCCAAUGUGCAAAACACAUCACCUUUUGCCAUUCGGCAAAGCCACUAUUGCCUUCUCGCCUCCGC------------------AAUCGCAAUGGCACGCACAAGCAAUUGAA ......(((((((((.((((((.........)))))))))).)))...((((...((((......((....))------------------....))))))))..)).(((....))).. ( -24.80) >DroSec_CAF1 8695 102 + 1 UAAUGGGUGCCAAUGUGCAAAACACAUCACCUUUCGCCAUUCGGCAAAGCCACUAUUGCCUUCUCGCCUCUGC------------------AAUCGCAAUGGCACGCACAAGCAAUUGAA ....(((((...(((((.....))))))))))...((.....(((((........))))).....(((..(((------------------....)))..)))..)).(((....))).. ( -27.30) >DroSim_CAF1 10053 102 + 1 UAAUGGGCGCCAAUGUGCAAAACACAUCACCUUUCGCCAUUCGGCAAAGCCACUAUUGCCUUCUCGCCUCCGC------------------AAUCGCAAUGGCACGCACAAGCAAUUGAA ......(((...(((((.....)))))........((((((((((((........))))).....((....))------------------....).)))))).))).(((....))).. ( -23.70) >DroYak_CAF1 5830 120 + 1 UAAUGGGCGCCAAUGUGCGAAACACAUCACCUUUCGCCAUUCGGCAAAGCCACUAUUGCCUUCUCGCCUCCGCCAUCGCCAUCGCCAUUGCAAUAGCAAUGGCACGCACAAGCAAUUGAA .....((((((((((.((((((.........)))))))))).)))...))).(.(((((......((....))....((....((((((((....))))))))..))....))))).).. ( -36.40) >consensus UAAUGGGCGCCAAUGUGCAAAACACAUCACCUUUCGCCAUUCGGCAAAGCCACUAUUGCCUUCUCGCCUCCGC__________________AAUCGCAAUGGCACGCACAAGCAAUUGAA ...((.(((...(((((.....)))))........((((((((((((........))))).....((....))......................).)))))).))).)).......... (-20.79 = -20.60 + -0.19)

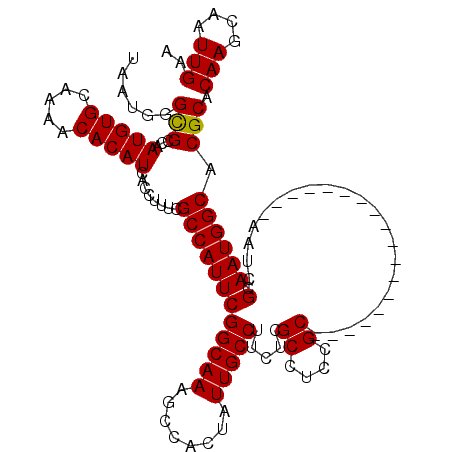
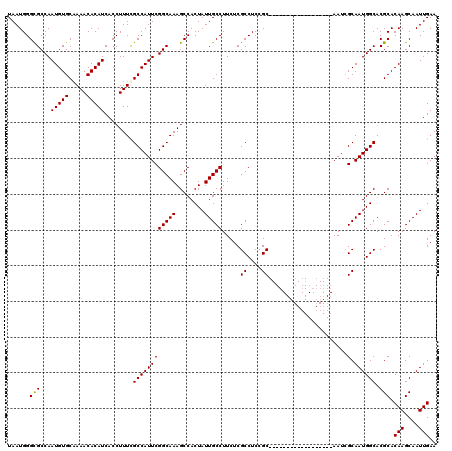
| Location | 6,266,488 – 6,266,590 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.64 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -33.70 |
| Energy contribution | -32.95 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6266488 102 - 22224390 UUCAAUUGCUUGUGCGUGCCAUUGCGAUU------------------GCGGAGGCGAGAAGGCAAUAGUGGCUUUGCCGAAUGGCAAAAGGUGAUGUGUUUUGCACAUUGGCGCCCAUUA ....(((((((...(.((((.((((....------------------)))).)))).).))))))).(.(((((((((....)))))).(.(((((((.....))))))).))))).... ( -35.70) >DroSec_CAF1 8695 102 - 1 UUCAAUUGCUUGUGCGUGCCAUUGCGAUU------------------GCAGAGGCGAGAAGGCAAUAGUGGCUUUGCCGAAUGGCGAAAGGUGAUGUGUUUUGCACAUUGGCACCCAUUA ....(((((((...(.((((.((((....------------------)))).)))).).)))))))(((((...((((.((((((((((.(.....).)))))).)))))))).))))). ( -35.20) >DroSim_CAF1 10053 102 - 1 UUCAAUUGCUUGUGCGUGCCAUUGCGAUU------------------GCGGAGGCGAGAAGGCAAUAGUGGCUUUGCCGAAUGGCGAAAGGUGAUGUGUUUUGCACAUUGGCGCCCAUUA ....(((((((...(.((((.((((....------------------)))).)))).).))))))).(.(((((((((....)))))).(.(((((((.....))))))).))))).... ( -35.40) >DroYak_CAF1 5830 120 - 1 UUCAAUUGCUUGUGCGUGCCAUUGCUAUUGCAAUGGCGAUGGCGAUGGCGGAGGCGAGAAGGCAAUAGUGGCUUUGCCGAAUGGCGAAAGGUGAUGUGUUUCGCACAUUGGCGCCCAUUA ....(((((((.(((.(((((((((((((((....)))))))))))))))...)))...)))))))(((((...((((.((((((((((.(.....).)))))).)))))))).))))). ( -53.30) >consensus UUCAAUUGCUUGUGCGUGCCAUUGCGAUU__________________GCGGAGGCGAGAAGGCAAUAGUGGCUUUGCCGAAUGGCGAAAGGUGAUGUGUUUUGCACAUUGGCGCCCAUUA ....(((((((...(.((((.((((......................)))).)))).).)))))))(((((...((((.((((((((((.(.....).)))))).)))))))).))))). (-33.70 = -32.95 + -0.75)

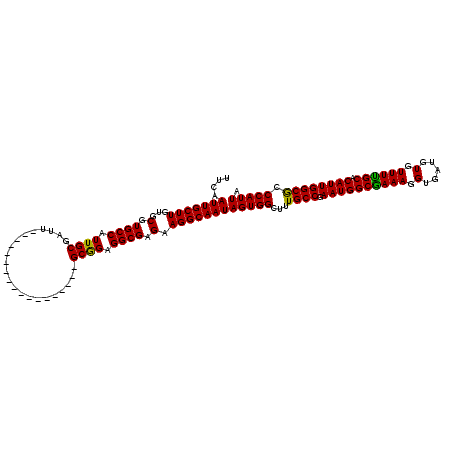
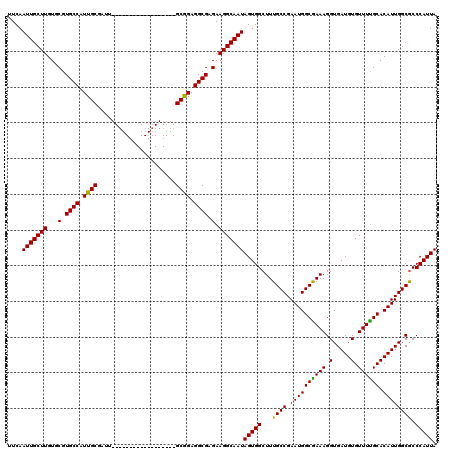
| Location | 6,266,528 – 6,266,630 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.09 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -21.23 |
| Energy contribution | -22.97 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6266528 102 + 22224390 UCGGCAAAGCCACUAUUGCCUUCUCGCCUCCGC------------------AAUCGCAAUGGCACGCACAAGCAAUUGAAAUUGUAUUAAAUUGAAUUUUAAUAUAUUGCUGGAGAAACA ..(((((........)))))((((((((...((------------------....))...)))..((((((....)))....(((((((((......))))))))).)))..)))))... ( -26.90) >DroSec_CAF1 8735 102 + 1 UCGGCAAAGCCACUAUUGCCUUCUCGCCUCUGC------------------AAUCGCAAUGGCACGCACAAGCAAUUGAAAUUGUAUUAAAUUGAAUUUUAAUAUAUUGCUGGAGAAACA ..(((((........)))))((((((((..(((------------------....)))..)))..((((((....)))....(((((((((......))))))))).)))..)))))... ( -29.30) >DroSim_CAF1 10093 102 + 1 UCGGCAAAGCCACUAUUGCCUUCUCGCCUCCGC------------------AAUCGCAAUGGCACGCACAAGCAAUUGAAAUUGUAUUAAAUUGAAUUUUAAUAUAUUGCCGGAGAAACA ..(((((........)))))((((((((...((------------------....))...)))..((((((....)))....(((((((((......))))))))).)))..)))))... ( -26.50) >DroYak_CAF1 5870 120 + 1 UCGGCAAAGCCACUAUUGCCUUCUCGCCUCCGCCAUCGCCAUCGCCAUUGCAAUAGCAAUGGCACGCACAAGCAAUUGAAAUUGUAUUAAAUUGAAUUUUAAUAUAUUGCUGGAGCAACA ..(((((........))))).....((.(((............((((((((....))))))))..((((((....)))....(((((((((......))))))))).))).))))).... ( -35.60) >consensus UCGGCAAAGCCACUAUUGCCUUCUCGCCUCCGC__________________AAUCGCAAUGGCACGCACAAGCAAUUGAAAUUGUAUUAAAUUGAAUUUUAAUAUAUUGCUGGAGAAACA ..(((((........)))))((((((((...((......................))...)))..((((((....)))....(((((((((......))))))))).)))..)))))... (-21.23 = -22.97 + 1.75)

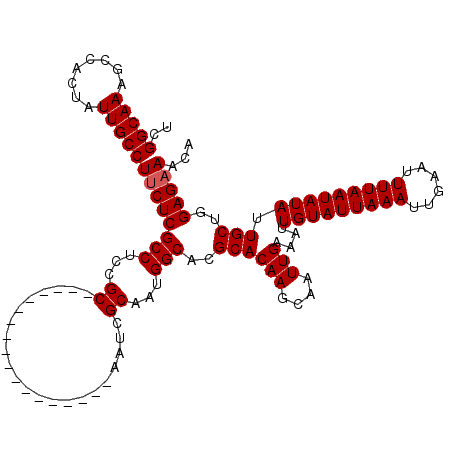
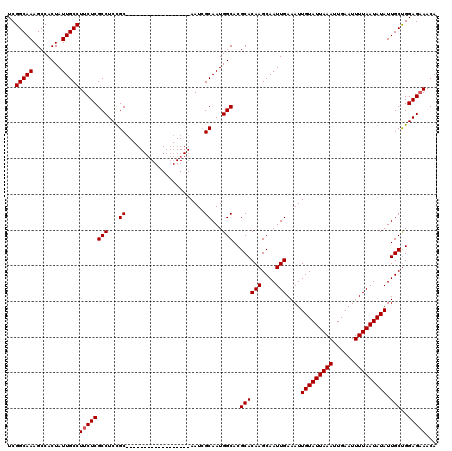
| Location | 6,266,528 – 6,266,630 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.09 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -22.14 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6266528 102 - 22224390 UGUUUCUCCAGCAAUAUAUUAAAAUUCAAUUUAAUACAAUUUCAAUUGCUUGUGCGUGCCAUUGCGAUU------------------GCGGAGGCGAGAAGGCAAUAGUGGCUUUGCCGA ...(((((..(((...(((((((......)))))))((((....))))....)))..(((.((((....------------------)))).))))))))(((((........))))).. ( -24.60) >DroSec_CAF1 8735 102 - 1 UGUUUCUCCAGCAAUAUAUUAAAAUUCAAUUUAAUACAAUUUCAAUUGCUUGUGCGUGCCAUUGCGAUU------------------GCAGAGGCGAGAAGGCAAUAGUGGCUUUGCCGA ...(((((..(((...(((((((......)))))))((((....))))....)))..(((.((((....------------------)))).))))))))(((((........))))).. ( -25.40) >DroSim_CAF1 10093 102 - 1 UGUUUCUCCGGCAAUAUAUUAAAAUUCAAUUUAAUACAAUUUCAAUUGCUUGUGCGUGCCAUUGCGAUU------------------GCGGAGGCGAGAAGGCAAUAGUGGCUUUGCCGA ........((((((..(((((((......)))))))........(((((((...(.((((.((((....------------------)))).)))).).))))))).......)))))). ( -27.00) >DroYak_CAF1 5870 120 - 1 UGUUGCUCCAGCAAUAUAUUAAAAUUCAAUUUAAUACAAUUUCAAUUGCUUGUGCGUGCCAUUGCUAUUGCAAUGGCGAUGGCGAUGGCGGAGGCGAGAAGGCAAUAGUGGCUUUGCCGA (.(((((((.(((...(((((((......)))))))((((....))))....)))..((((((((((((((....))))))))))))))))).)))).).(((((........))))).. ( -43.70) >consensus UGUUUCUCCAGCAAUAUAUUAAAAUUCAAUUUAAUACAAUUUCAAUUGCUUGUGCGUGCCAUUGCGAUU__________________GCGGAGGCGAGAAGGCAAUAGUGGCUUUGCCGA ...(((((..(((...(((((((......)))))))((((....))))....)))..(((.((((......................)))).))))))))(((((........))))).. (-22.14 = -22.20 + 0.06)

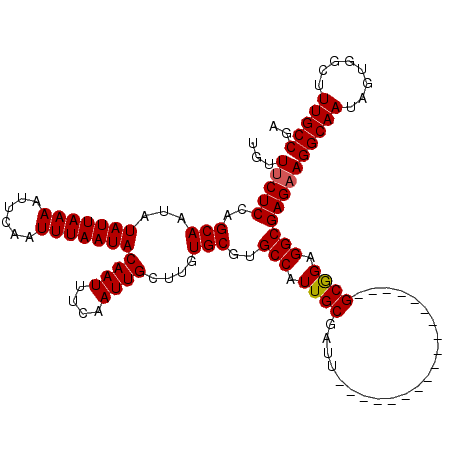

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:02 2006