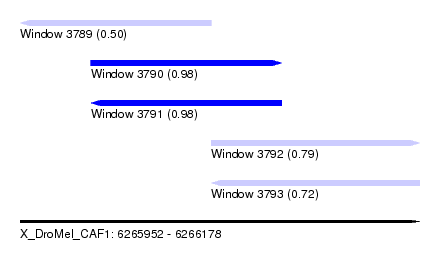
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,265,952 – 6,266,178 |
| Length | 226 |
| Max. P | 0.982093 |

| Location | 6,265,952 – 6,266,060 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.23 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -19.37 |
| Energy contribution | -20.03 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6265952 108 - 22224390 UUGUUUGAAGUGCGUGUUGCAUCGUUCAACGCUGGCGUUAUUUAUGCAGCAACAGCAAUAGCAACAGCAACAUGAACGGAAGCGGCAACAACGUCUGCCAGUCGAUCU (((.(((.((.(((((((((.(((((((..((((..(((((...(((.......))))))))..))))....))))))).....))))).))))))..))).)))... ( -30.90) >DroSec_CAF1 8132 108 - 1 UUGUUUGAAGUGCGUGUUGCAUCGUUCAACGCUGGCGUUAUUUAUGCAGAAACAGCAAUAGCAACAGCAACAUGAACGGAAGCGGCAACAACGUCUGCCAGUCGAACU ..((((((.(((((((((((.(((((((..((((..(((((...(((.......))))))))..))))....))))))).....))))).))))..))...)))))). ( -33.90) >DroSim_CAF1 9490 108 - 1 UUGUUUGAAGUGCGUGUUGCAUCGUUCAACGCUGGCGUUAUUUAUGCAGCAACAGCAAUAGCAACAGCAAGANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN (((((((...(((.((((((((.....((((....))))....))))))))...)))....)))..))))...................................... ( -20.00) >consensus UUGUUUGAAGUGCGUGUUGCAUCGUUCAACGCUGGCGUUAUUUAUGCAGCAACAGCAAUAGCAACAGCAACAUGAACGGAAGCGGCAACAACGUCUGCCAGUCGA_CU (((((((...(((.((((((((.....((((....))))....))))))))...)))....)))..)))).............((((........))))......... (-19.37 = -20.03 + 0.67)

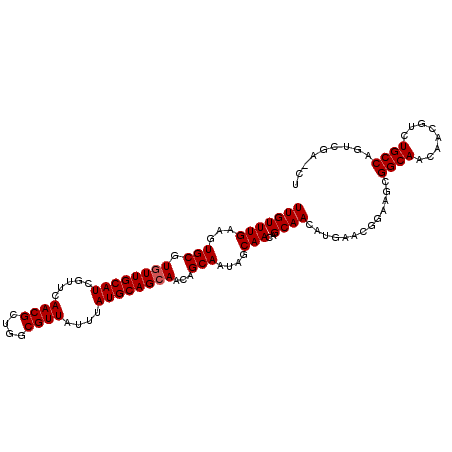
| Location | 6,265,992 – 6,266,100 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 97.52 |
| Mean single sequence MFE | -26.81 |
| Consensus MFE | -25.27 |
| Energy contribution | -25.27 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.85 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6265992 108 + 22224390 GCUGUUGCUAUUGCUGUUGCUGCAUAAAUAACGCCAGCGUUGAACGAUGCAACACGCACUUCAAACAACAAUAAAACAAGCAACAGCAACGCCCACAACAACAGAACU .((((((...(((((((((((.......(((((....)))))...(((((.....)))..))................))))))))))).(....)..)))))).... ( -27.90) >DroSec_CAF1 8172 107 + 1 GCUGUUGCUAUUGCUGUUUCUGCAUAAAUAACGCCAGCGUUGAACGAUGCAACACGCACUUCAAACAACAAUAACACAAGCAACAGCA-CACCCACAACAACAGAACU (((((((((((((.(((((.(((((...(((((....)))))....)))))...........))))).))))......))))))))).-................... ( -25.60) >DroSim_CAF1 9530 107 + 1 GCUGUUGCUAUUGCUGUUGCUGCAUAAAUAACGCCAGCGUUGAACGAUGCAACACGCACUUCAAACAACAAUAACACAAGCAACAGCA-CACCCACAACAACAGAACU .((((((....((((((((((.......(((((....)))))...(((((.....)))..))................))))))))))-.........)))))).... ( -26.92) >consensus GCUGUUGCUAUUGCUGUUGCUGCAUAAAUAACGCCAGCGUUGAACGAUGCAACACGCACUUCAAACAACAAUAACACAAGCAACAGCA_CACCCACAACAACAGAACU (((((((((((((.((((..(((((...(((((....)))))....)))))............)))).))))......)))))))))..................... (-25.27 = -25.27 + 0.00)



| Location | 6,265,992 – 6,266,100 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 97.52 |
| Mean single sequence MFE | -35.43 |
| Consensus MFE | -34.53 |
| Energy contribution | -34.87 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6265992 108 - 22224390 AGUUCUGUUGUUGUGGGCGUUGCUGUUGCUUGUUUUAUUGUUGUUUGAAGUGCGUGUUGCAUCGUUCAACGCUGGCGUUAUUUAUGCAGCAACAGCAAUAGCAACAGC ......((((((((....((((((((((((.........((((..(((..(((.....))))))..))))....((((.....)))))))))))))))).)))))))) ( -38.30) >DroSec_CAF1 8172 107 - 1 AGUUCUGUUGUUGUGGGUG-UGCUGUUGCUUGUGUUAUUGUUGUUUGAAGUGCGUGUUGCAUCGUUCAACGCUGGCGUUAUUUAUGCAGAAACAGCAAUAGCAACAGC ....((((((((((.....-(((((((..((((((((.(((((..(((..(((.....))))))..))))).)))).........)))).))))))))))))))))). ( -30.90) >DroSim_CAF1 9530 107 - 1 AGUUCUGUUGUUGUGGGUG-UGCUGUUGCUUGUGUUAUUGUUGUUUGAAGUGCGUGUUGCAUCGUUCAACGCUGGCGUUAUUUAUGCAGCAACAGCAAUAGCAACAGC ....((((((((((.....-((((((((((.........((((..(((..(((.....))))))..))))....((((.....)))))))))))))))))))))))). ( -37.10) >consensus AGUUCUGUUGUUGUGGGUG_UGCUGUUGCUUGUGUUAUUGUUGUUUGAAGUGCGUGUUGCAUCGUUCAACGCUGGCGUUAUUUAUGCAGCAACAGCAAUAGCAACAGC ....((((((((((......((((((((((.........((((..(((..(((.....))))))..))))....((((.....)))))))))))))))))))))))). (-34.53 = -34.87 + 0.33)



| Location | 6,266,060 – 6,266,178 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.88 |
| Mean single sequence MFE | -24.26 |
| Consensus MFE | -21.14 |
| Energy contribution | -21.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6266060 118 + 22224390 CAAUAAAACAAGCAACAGCAACGCCCACAACAACAGAACUUUGCACGUUUUGCAUACAAAGCAGAAUAACCCGCAAAAAAAAAGUAUCUGGCGGGUGAGGG--AGCCGGCAAGAAAUAUU .................((....(((....................(((((((.......))))))).((((((................))))))..)))--.....)).......... ( -23.79) >DroSec_CAF1 8240 114 + 1 CAAUAACACAAGCAACAGCA-CACCCACAACAACAGAACUUUGCACGUUUUGCAUACAAAGCAGAAUAACCCGCAAA---AAAGUAUCUGGCGGGUGAGGG--AGCCAGCAAGAAAUAUU ...........((....((.-..(((....................(((((((.......))))))).((((((...---..........))))))..)))--.))..)).......... ( -22.42) >DroSim_CAF1 9598 114 + 1 CAAUAACACAAGCAACAGCA-CACCCACAACAACAGAACUUUGCACGUUUUGCAUACAAAGCAGAAUAACCCGCAAA---AAAGUAUCUGGCGGGUGAGGG--AGCCAGCAAGAAAUAUU ...........((....((.-..(((....................(((((((.......))))))).((((((...---..........))))))..)))--.))..)).......... ( -22.42) >DroYak_CAF1 5380 116 + 1 CACUAACAGUAGCAACAGCA-CACCCACAACAACAAAACGUUGCACGUUUUGCAUACAAAGCAGAAUAACCCGCAAA---AAAGUAUCUGGCGGGUGAGGGGAAAGCUGCAAGAAAUAUU ..((..((((.((....)).-..(((....((((.....))))...(((((((.......))))))).((((((...---..........))))))..)))....))))..))....... ( -28.42) >consensus CAAUAACACAAGCAACAGCA_CACCCACAACAACAGAACUUUGCACGUUUUGCAUACAAAGCAGAAUAACCCGCAAA___AAAGUAUCUGGCGGGUGAGGG__AGCCAGCAAGAAAUAUU ...........((....))....(((....................(((((((.......))))))).((((((................))))))..)))................... (-21.14 = -21.14 + 0.00)

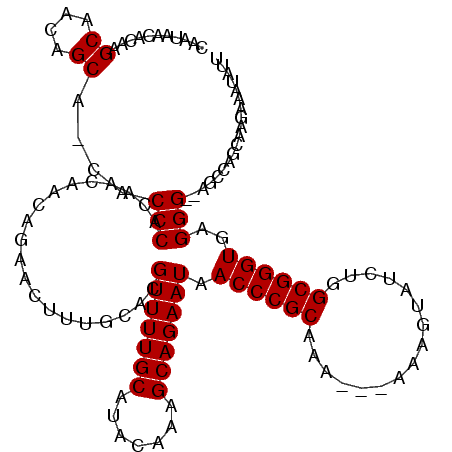
| Location | 6,266,060 – 6,266,178 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.88 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -25.39 |
| Energy contribution | -25.70 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6266060 118 - 22224390 AAUAUUUCUUGCCGGCU--CCCUCACCCGCCAGAUACUUUUUUUUUGCGGGUUAUUCUGCUUUGUAUGCAAAACGUGCAAAGUUCUGUUGUUGUGGGCGUUGCUGUUGCUUGUUUUAUUG ((((...(..((((((.--(((.(((((((.(((........))).))))))......((((((((((.....)))))))))).........).)))....))))..))..)...)))). ( -32.40) >DroSec_CAF1 8240 114 - 1 AAUAUUUCUUGCUGGCU--CCCUCACCCGCCAGAUACUUU---UUUGCGGGUUAUUCUGCUUUGUAUGCAAAACGUGCAAAGUUCUGUUGUUGUGGGUG-UGCUGUUGCUUGUGUUAUUG ((((...(..((.(((.--(((.(((((((.(((.....)---)).))))))......((((((((((.....)))))))))).........).)))..-.)))...))..)...)))). ( -32.40) >DroSim_CAF1 9598 114 - 1 AAUAUUUCUUGCUGGCU--CCCUCACCCGCCAGAUACUUU---UUUGCGGGUUAUUCUGCUUUGUAUGCAAAACGUGCAAAGUUCUGUUGUUGUGGGUG-UGCUGUUGCUUGUGUUAUUG ((((...(..((.(((.--(((.(((((((.(((.....)---)).))))))......((((((((((.....)))))))))).........).)))..-.)))...))..)...)))). ( -32.40) >DroYak_CAF1 5380 116 - 1 AAUAUUUCUUGCAGCUUUCCCCUCACCCGCCAGAUACUUU---UUUGCGGGUUAUUCUGCUUUGUAUGCAAAACGUGCAACGUUUUGUUGUUGUGGGUG-UGCUGUUGCUACUGUUAGUG ..........(((((........(((((((...((((...---...(((((....)))))...))))((((((((.....))))))))....)))))))-.))))).((((....)))). ( -32.40) >consensus AAUAUUUCUUGCUGGCU__CCCUCACCCGCCAGAUACUUU___UUUGCGGGUUAUUCUGCUUUGUAUGCAAAACGUGCAAAGUUCUGUUGUUGUGGGUG_UGCUGUUGCUUGUGUUAUUG ..........((.(((...(((.(((((((.(((.........)))))))))......((((((((((.....)))))))))).........).)))....)))...))........... (-25.39 = -25.70 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:58 2006