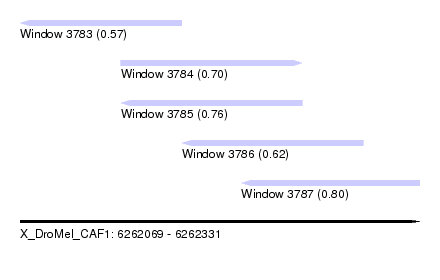
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,262,069 – 6,262,331 |
| Length | 262 |
| Max. P | 0.800335 |

| Location | 6,262,069 – 6,262,175 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.04 |
| Mean single sequence MFE | -24.14 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.30 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6262069 106 - 22224390 AAGCUCUGGAAAAAGUGCAGAACAUAAAAAUGUUUGCUUCCGGCCAACAAUACAGAAGCCGCAAUAUUCACACUUUUACAGCUGACUGGGCAC----ACACAUCCAGGCA---------- ..((.((((((((((((..((((((....))))))(((((.(..........).)))))...........)))))))...(((.....)))..----.....))))))).---------- ( -26.80) >DroSec_CAF1 3734 97 - 1 AAGCUCUGGAAAAAGUGCAGAGCAUAAAAAUGUUUGCUUCCGGCCAACAAUACAGAAGCCGCAAUAUUCACACUUUUACAGCUGACUGGGCAC----ACAC------------------- ..((((((.(.....).)))))).......(((.((((..((((.............)))).................(((....))))))).----))).------------------- ( -21.12) >DroSim_CAF1 3668 105 - 1 AAGCUCUGGAAAAAGUGCAGAGCAUAAAAAUGUUUGCUUCCGGCCAACAAUACAGAAGCCGCAAUAUUCACACUUUUACAGCUGACUGGGCAC----ACACAUCCCGAC----------- ..((((((.(.....).)))))).......(((.((((..((((.............)))).................(((....))))))).----))).........----------- ( -21.12) >DroYak_CAF1 3205 120 - 1 AAGCUCUGGAAAAAGUGCAGAACAUAAAAAUGUUUGCUUCCGGCCAACAAUACAGAAGCCGCAAUAUUCACACUUUUACAGCUGACUGGGCAGACGCACACAUCCAGGCAUAUUCAGACA ..((.(((((....((((.((((((....))))))(((..((((.............)))).................(((....))))))....))))...)))))))........... ( -27.52) >consensus AAGCUCUGGAAAAAGUGCAGAACAUAAAAAUGUUUGCUUCCGGCCAACAAUACAGAAGCCGCAAUAUUCACACUUUUACAGCUGACUGGGCAC____ACACAUCCAGGC___________ ..((((.((.(((((((..((((((....))))))(((((.(..........).)))))...........)))))))....))....))))............................. (-18.55 = -18.30 + -0.25)

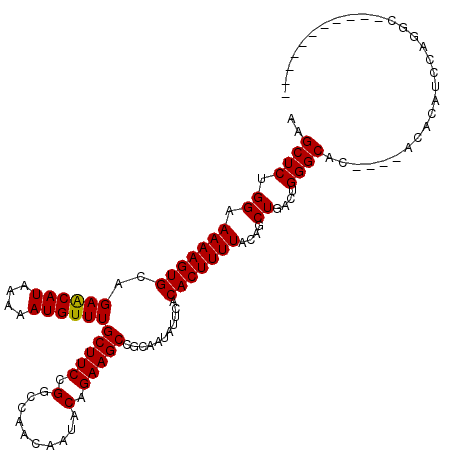
| Location | 6,262,135 – 6,262,254 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.54 |
| Mean single sequence MFE | -30.85 |
| Consensus MFE | -24.62 |
| Energy contribution | -25.12 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6262135 119 + 22224390 GAAGCAAACAUUUUUAUGUUCUGCACUUUUUCCAGAGCUUCUUCAUCUUCGCUCCAUACCCCUAUAU-AUCGUAAGGACGAAAAGGACGAGGUAGUUGGGCAUUUGAUUAAGCCCGGCUC ...((((((((....))))).)))(((((.(((.(((....)))...((((.(((.(((........-...))).)))))))..))).)))))((((((((..........)))))))). ( -31.70) >DroSec_CAF1 3791 120 + 1 GAAGCAAACAUUUUUAUGCUCUGCACUUUUUCCAGAGCUUCUUCAUCUUCGCUCCAUACCAGUAUACCGCCCUAAGGACGAAAAGGACCAUCUAGCUGAGCAUUUGAUUAAGCCCGGCUC ..(((......((..((((((.((......(((.((((............)))).............((((....)).))....))).......)).))))))..)).........))). ( -21.08) >DroSim_CAF1 3733 120 + 1 GAAGCAAACAUUUUUAUGCUCUGCACUUUUUCCAGAGCUUCUUCGUCUUCGCUCCAUACCAGUAUACCGCCCUAAGGACGAAAGGGACGAGGUAGCUGGGCAUUUGAUUAAGCCCGGCUC .................((((((.........))))))..(((((((((((.(((.((...((.....))..)).))))))...)))))))).((((((((..........)))))))). ( -35.40) >DroYak_CAF1 3285 108 + 1 GAAGCAAACAUUUUUAUGUUCUGCACUUUUUCCAGAGCUUCUUCAUCUUU-CUCC----CCCCAA-------AAGGGACAAAAAGGAUGAAGUAGCUGGGCAUUUGAUUAAGCCCGGUUC ...((((((((....))))).))).......((..((((.((((((((((-.(((----(.....-------..))))....)))))))))).))))((((..........))))))... ( -35.20) >consensus GAAGCAAACAUUUUUAUGCUCUGCACUUUUUCCAGAGCUUCUUCAUCUUCGCUCCAUACCACUAUAC_GCCCUAAGGACGAAAAGGACGAGGUAGCUGGGCAUUUGAUUAAGCCCGGCUC .................((((((.........))))))..(((((((((((.(((....................))))))...)))))))).((((((((..........)))))))). (-24.62 = -25.12 + 0.50)



| Location | 6,262,135 – 6,262,254 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.54 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -23.61 |
| Energy contribution | -23.43 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6262135 119 - 22224390 GAGCCGGGCUUAAUCAAAUGCCCAACUACCUCGUCCUUUUCGUCCUUACGAU-AUAUAGGGGUAUGGAGCGAAGAUGAAGAAGCUCUGGAAAAAGUGCAGAACAUAAAAAUGUUUGCUUC ...((((((((..(((..((((((..((((((.......((((....)))).-.....))))))))).)))....)))..)))).)))).......(((((.(((....))))))))... ( -33.02) >DroSec_CAF1 3791 120 - 1 GAGCCGGGCUUAAUCAAAUGCUCAGCUAGAUGGUCCUUUUCGUCCUUAGGGCGGUAUACUGGUAUGGAGCGAAGAUGAAGAAGCUCUGGAAAAAGUGCAGAGCAUAAAAAUGUUUGCUUC ...((((((((..(((..(((((.(((((((.(((((..........))))).))...)))))...)))))....)))..)))).)))).......(((((.(((....))))))))... ( -34.60) >DroSim_CAF1 3733 120 - 1 GAGCCGGGCUUAAUCAAAUGCCCAGCUACCUCGUCCCUUUCGUCCUUAGGGCGGUAUACUGGUAUGGAGCGAAGACGAAGAAGCUCUGGAAAAAGUGCAGAGCAUAAAAAUGUUUGCUUC .(((.((((..........)))).))).(.(((((...(((((((....(.(((....))).)..))).))))))))).)..((((((.(.....).))))))................. ( -35.80) >DroYak_CAF1 3285 108 - 1 GAACCGGGCUUAAUCAAAUGCCCAGCUACUUCAUCCUUUUUGUCCCUU-------UUGGGG----GGAG-AAAGAUGAAGAAGCUCUGGAAAAAGUGCAGAACAUAAAAAUGUUUGCUUC ...((((((..........))))((((.(((((((.(((((.((((..-------..))))----.)))-)).))))))).))))..)).......(((((.(((....))))))))... ( -39.90) >consensus GAGCCGGGCUUAAUCAAAUGCCCAGCUACCUCGUCCUUUUCGUCCUUAGGGC_GUAUACGGGUAUGGAGCGAAGAUGAAGAAGCUCUGGAAAAAGUGCAGAACAUAAAAAUGUUUGCUUC .(((.((((..........)))).)))...(((((...(((((((....................))).))))))))).(((((((((.(.....).))))((((....))))..))))) (-23.61 = -23.43 + -0.19)


| Location | 6,262,175 – 6,262,294 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.67 |
| Mean single sequence MFE | -39.55 |
| Consensus MFE | -29.12 |
| Energy contribution | -28.50 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6262175 119 - 22224390 GCCGAGAAUCUCAGUUUCAGUGCAAUUAGCUGCGAGCUGGGAGCCGGGCUUAAUCAAAUGCCCAACUACCUCGUCCUUUUCGUCCUUACGAU-AUAUAGGGGUAUGGAGCGAAGAUGAAG ..((((..(((((((((((((.......)))).)))))))))...((((..........))))......))))((.(((((((((.(((...-........))).))).)))))).)).. ( -37.50) >DroSec_CAF1 3831 120 - 1 GCCGAGAAUCUCAGUUUCAGUGCAAUUAGCUGCGAGCUGGGAGCCGGGCUUAAUCAAAUGCUCAGCUAGAUGGUCCUUUUCGUCCUUAGGGCGGUAUACUGGUAUGGAGCGAAGAUGAAG (((..(..(((((((((((((.......)))).)))))))))..).)))...(((...(((((.(((((((.(((((..........))))).))...)))))...)))))..))).... ( -38.50) >DroSim_CAF1 3773 120 - 1 GCCGAGAAUCUCAGUUUCAGUGCAAUUAGCUGCGAGCUGGGAGCCGGGCUUAAUCAAAUGCCCAGCUACCUCGUCCCUUUCGUCCUUAGGGCGGUAUACUGGUAUGGAGCGAAGACGAAG ...(((..(((((((((((((.......)))).)))))))))((.((((..........)))).))...)))(((...(((((((....(.(((....))).)..))).))))))).... ( -39.30) >DroYak_CAF1 3325 108 - 1 GCCGAGAAUCUCAGUUUCAGUGCAAUUAGCUGCAAGCUGGGAACCGGGCUUAAUCAAAUGCCCAGCUACUUCAUCCUUUUUGUCCCUU-------UUGGGG----GGAG-AAAGAUGAAG ((......(((((((((((((.......)))).)))))))))...((((..........)))).))..(((((((.(((((.((((..-------..))))----.)))-)).))))))) ( -42.90) >consensus GCCGAGAAUCUCAGUUUCAGUGCAAUUAGCUGCGAGCUGGGAGCCGGGCUUAAUCAAAUGCCCAGCUACCUCGUCCUUUUCGUCCUUAGGGC_GUAUACGGGUAUGGAGCGAAGAUGAAG .....(..(((((((((((((.......)))).)))))))))..)((((..........)))).......(((((...(((((((....................))).))))))))).. (-29.12 = -28.50 + -0.62)

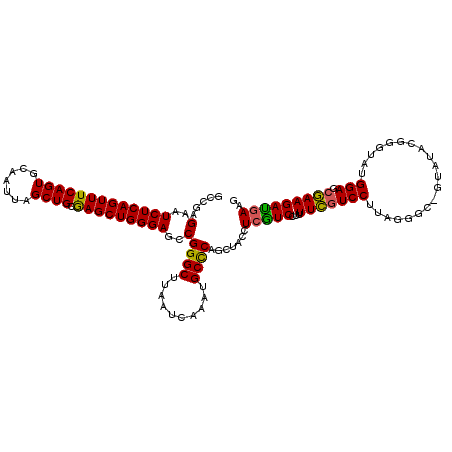

| Location | 6,262,214 – 6,262,331 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.55 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -32.11 |
| Energy contribution | -32.30 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6262214 117 - 22224390 AACCGG---GCACUGGCAGCACACUUUGCAUUCCAGCAAUGCCGAGAAUCUCAGUUUCAGUGCAAUUAGCUGCGAGCUGGGAGCCGGGCUUAAUCAAAUGCCCAACUACCUCGUCCUUUU ....((---((...(((......(((.(((((.....))))).)))..(((((((((((((.......)))).))))))))))))((((..........)))).........)))).... ( -39.80) >DroSec_CAF1 3871 117 - 1 AACCGG---GCACUGGCAGCACACUUUGCAUUCCGGCAAUGCCGAGAAUCUCAGUUUCAGUGCAAUUAGCUGCGAGCUGGGAGCCGGGCUUAAUCAAAUGCUCAGCUAGAUGGUCCUUUU ....((---((.(((((.((..........(((((((...)))).)))(((((((((((((.......)))).))))))))))).((((..........)))).)))))...)))).... ( -40.40) >DroSim_CAF1 3813 117 - 1 AACCGG---GCACUGGCAGCACACUUUGCAUUCCGGCAAUGCCGAGAAUCUCAGUUUCAGUGCAAUUAGCUGCGAGCUGGGAGCCGGGCUUAAUCAAAUGCCCAGCUACCUCGUCCCUUU ....((---((.((.(((((.....(((((((.((((...)))).(((.......))))))))))...))))).))..((((((.((((..........)))).))).))).)))).... ( -42.90) >DroYak_CAF1 3353 120 - 1 AACCAGGCGGCAAUGGGAGCACACUUUGCAUUCUGGCAAUGCCGAGAAUCUCAGUUUCAGUGCAAUUAGCUGCAAGCUGGGAACCGGGCUUAAUCAAAUGCCCAGCUACUUCAUCCUUUU ....((((((((.((((((((.....))).)))))....))))).((((((((((((((((.......)))).)))))))))...((((..........))))......)))..)))... ( -36.40) >consensus AACCGG___GCACUGGCAGCACACUUUGCAUUCCGGCAAUGCCGAGAAUCUCAGUUUCAGUGCAAUUAGCUGCGAGCUGGGAGCCGGGCUUAAUCAAAUGCCCAGCUACCUCGUCCUUUU .........((...(((.(((.....))).(((((((...)))).)))(((((((((((((.......)))).))))))))))))((((..........)))).)).............. (-32.11 = -32.30 + 0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:53 2006