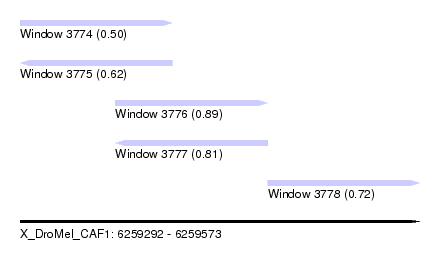
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,259,292 – 6,259,573 |
| Length | 281 |
| Max. P | 0.886657 |

| Location | 6,259,292 – 6,259,399 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -23.20 |
| Energy contribution | -23.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 1.00 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6259292 107 + 22224390 UUUUUUUUUUUUUGUUUUUUUGUCACUUCACUGGAAGCGCGCUCAAAGGAGGCUAAAUAUUGCAUUACGUAUCCUUGCACUUUAUGGCUGCUCCUUGGGCUCUAGUU .............................((((((.....((((((.((((((((.....((((...........)))).....))))..)))))))))))))))). ( -23.20) >DroSec_CAF1 941 107 + 1 CUAUUUUUUUUUUGUUUUUUUGUCACUUCACUGGAAGCGCGCUCAAAGGAGGCUAAAUAUUGCAUUACGUAUCCUUGCACUUUAUGGCUGCUCCUUGGGCUCUAGUU .............................((((((.....((((((.((((((((.....((((...........)))).....))))..)))))))))))))))). ( -23.20) >DroSim_CAF1 945 107 + 1 CUAUUUUUUUUUUGUUUUUUUGUCACUUCACUGGAAGCGCGCUCAAAGGAGGCUAAAUAUUGCAUUACGUAUCCUUGCACUUUAUGGCUGCUCCUUGGGCUCUAGUU .............................((((((.....((((((.((((((((.....((((...........)))).....))))..)))))))))))))))). ( -23.20) >consensus CUAUUUUUUUUUUGUUUUUUUGUCACUUCACUGGAAGCGCGCUCAAAGGAGGCUAAAUAUUGCAUUACGUAUCCUUGCACUUUAUGGCUGCUCCUUGGGCUCUAGUU .............................((((((.....((((((.((((((((.....((((...........)))).....))))..)))))))))))))))). (-23.20 = -23.20 + -0.00)


| Location | 6,259,292 – 6,259,399 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -20.60 |
| Energy contribution | -20.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6259292 107 - 22224390 AACUAGAGCCCAAGGAGCAGCCAUAAAGUGCAAGGAUACGUAAUGCAAUAUUUAGCCUCCUUUGAGCGCGCUUCCAGUGAAGUGACAAAAAAACAAAAAAAAAAAAA .......(((.((((((..((..((((.((((.(....)....))))...)))))))))))).).))(((((((....)))))).)..................... ( -20.60) >DroSec_CAF1 941 107 - 1 AACUAGAGCCCAAGGAGCAGCCAUAAAGUGCAAGGAUACGUAAUGCAAUAUUUAGCCUCCUUUGAGCGCGCUUCCAGUGAAGUGACAAAAAAACAAAAAAAAAAUAG .......(((.((((((..((..((((.((((.(....)....))))...)))))))))))).).))(((((((....)))))).)..................... ( -20.60) >DroSim_CAF1 945 107 - 1 AACUAGAGCCCAAGGAGCAGCCAUAAAGUGCAAGGAUACGUAAUGCAAUAUUUAGCCUCCUUUGAGCGCGCUUCCAGUGAAGUGACAAAAAAACAAAAAAAAAAUAG .......(((.((((((..((..((((.((((.(....)....))))...)))))))))))).).))(((((((....)))))).)..................... ( -20.60) >consensus AACUAGAGCCCAAGGAGCAGCCAUAAAGUGCAAGGAUACGUAAUGCAAUAUUUAGCCUCCUUUGAGCGCGCUUCCAGUGAAGUGACAAAAAAACAAAAAAAAAAUAG .......(((.((((((..((..((((.((((.(....)....))))...)))))))))))).).))(((((((....)))))).)..................... (-20.60 = -20.60 + -0.00)

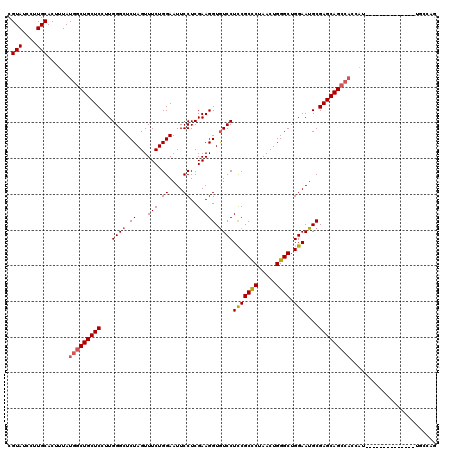
| Location | 6,259,359 – 6,259,466 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.32 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -32.42 |
| Energy contribution | -33.05 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6259359 107 + 22224390 CGUAUCCUUGCACUUUAUGGCUGCUCCUUGGGCUCUAGUUUCUGGAAUUCCUCGAAGGUGUCCUUCGCCCUAACUGGGCUGGAAUGCGAGCAGCCACCAUU-------------UGCCAG ......((.(((..(..(((((((((.......(((((...)))))(((((..(((((...)))))((((.....)))).)))))..)))))))))..)..-------------))).)) ( -40.30) >DroSec_CAF1 1008 106 + 1 CGUAUCCUUGCACUUUAUGGCUGCUCCUUGGGCUCUAGUUUCUGGAAUUCCUCGAAGGUGUCCUCCGCCCUAACUGGGCUGGAAUGCGAGCAGCCACCAU--------------UGCCAG ......((.(((.....(((((((((...((((.((...(((.((....))..))))).))))(((((((.....)))).)))....)))))))))....--------------))).)) ( -39.10) >DroSim_CAF1 1012 93 + 1 CGUAUCCUUGCACUUUAUGGCUGCUCCUUGGGCUCUAGUUUCUGGAAUUCCUCGAAGGUGUCCUCCGCCCUAACUGGGCUGGAAUGCGAGCAG--------------------------- .(((....))).........((((((...((((.((...(((.((....))..))))).))))(((((((.....)))).)))....))))))--------------------------- ( -29.00) >DroYak_CAF1 546 120 + 1 CGUAUCCUUGCACUUUAUGGCUGCUCCUUGGGCUCUAGUUUCUGGAAUUCCUCGAAGGUAUCCUCCGCUCUAACUGGGCUGGAAUGCGAGCAGCCACCAUUACCAGCUACCAGCUACCAG .(((....)))......(((((((((((((((.(((((...)))))...)))..)))((((..(((((((.....)))).))))))))))))))))........(((.....)))..... ( -35.70) >consensus CGUAUCCUUGCACUUUAUGGCUGCUCCUUGGGCUCUAGUUUCUGGAAUUCCUCGAAGGUGUCCUCCGCCCUAACUGGGCUGGAAUGCGAGCAGCCACCAU______________UGCCAG .(((....)))......(((((((((...((((.((...(((.((....))..))))).))))(((((((.....)))).)))....)))))))))........................ (-32.42 = -33.05 + 0.63)


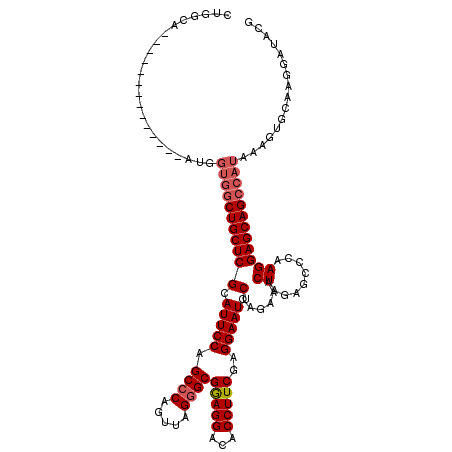
| Location | 6,259,359 – 6,259,466 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.32 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -30.69 |
| Energy contribution | -31.75 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6259359 107 - 22224390 CUGGCA-------------AAUGGUGGCUGCUCGCAUUCCAGCCCAGUUAGGGCGAAGGACACCUUCGAGGAAUUCCAGAAACUAGAGCCCAAGGAGCAGCCAUAAAGUGCAAGGAUACG ((.(((-------------....(((((((((((.(((((.((((.....))))(((((...)))))..))))).)......((........))))))))))))....))).))...... ( -42.30) >DroSec_CAF1 1008 106 - 1 CUGGCA--------------AUGGUGGCUGCUCGCAUUCCAGCCCAGUUAGGGCGGAGGACACCUUCGAGGAAUUCCAGAAACUAGAGCCCAAGGAGCAGCCAUAAAGUGCAAGGAUACG ((.(((--------------...(((((((((((.(((((.((((.....))))(((((...)))))..))))).)......((........))))))))))))....))).))...... ( -41.40) >DroSim_CAF1 1012 93 - 1 ---------------------------CUGCUCGCAUUCCAGCCCAGUUAGGGCGGAGGACACCUUCGAGGAAUUCCAGAAACUAGAGCCCAAGGAGCAGCCAUAAAGUGCAAGGAUACG ---------------------------(((((((.(((((.((((.....))))(((((...)))))..))))).)......((........)))))))).................... ( -25.70) >DroYak_CAF1 546 120 - 1 CUGGUAGCUGGUAGCUGGUAAUGGUGGCUGCUCGCAUUCCAGCCCAGUUAGAGCGGAGGAUACCUUCGAGGAAUUCCAGAAACUAGAGCCCAAGGAGCAGCCAUAAAGUGCAAGGAUACG ....((((.....))))(((...((((((((((((......))...(((..((((((((...)))))).((....)).....))..))).....))))))))))....)))......... ( -36.10) >consensus CUGGCA______________AUGGUGGCUGCUCGCAUUCCAGCCCAGUUAGGGCGGAGGACACCUUCGAGGAAUUCCAGAAACUAGAGCCCAAGGAGCAGCCAUAAAGUGCAAGGAUACG .......................(((((((((((.(((((.((((.....))))(((((...)))))..))))).)......((........))))))))))))................ (-30.69 = -31.75 + 1.06)

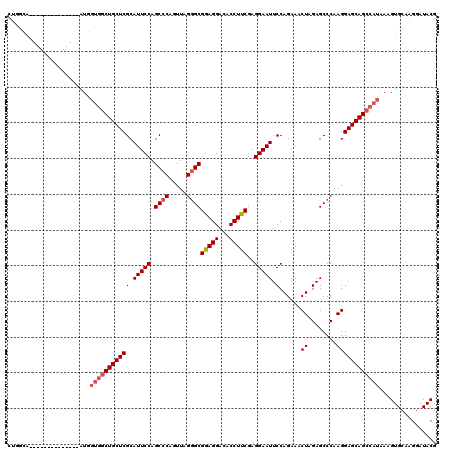
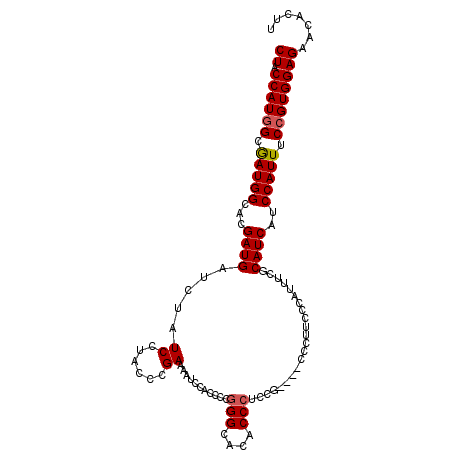
| Location | 6,259,466 – 6,259,573 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 89.59 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -17.59 |
| Energy contribution | -18.03 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6259466 107 + 22224390 CUACCAUGGCGAUGGCACGAUGAUCUAUCCUACCCGAAAAUCCAACCCUGGCACACCCCGUGCUGUCCUUUCCCAUUUCGCAUCAUCCAUUUCCGUGGAGAACACUU ((.((((((.(((((...((((............(((((..........(((((.....)))))...........)))))))))..))))).))))))))....... ( -25.30) >DroSec_CAF1 1114 102 + 1 CUACCAUGGCGAUGGCACGAUG-UCUAUCCCACCCGAAAAUCCACCCCGGGCACCCCCUCCG----CCCUUCCCAUUUCACAUCAUCCAUUUCCGUGGAGAACACUU ((.((((((.(((((...((((-(...((......))...........((((.........)----)))..........)))))..))))).))))))))....... ( -25.90) >DroSim_CAF1 1105 103 + 1 CUACCAUAGCAAUGGCACGAUGAUCUAUCCUACCCGAAAAUCCACCCCGGGCACUCCCUCCG----CCCUUCCCAUUUCGCAUCAUCCAUUUCCGUGGAGAACACUU ((.(((..(.(((((...((((.....((......))...........((((.........)----)))...........))))..))))).)..)))))....... ( -19.10) >consensus CUACCAUGGCGAUGGCACGAUGAUCUAUCCUACCCGAAAAUCCACCCCGGGCACACCCUCCG____CCCUUCCCAUUUCGCAUCAUCCAUUUCCGUGGAGAACACUU ((.((((((.(((((...((((.....((......))...........(((....)))......................))))..))))).))))))))....... (-17.59 = -18.03 + 0.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:44 2006