
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,259,067 – 6,259,252 |
| Length | 185 |
| Max. P | 0.945432 |

| Location | 6,259,067 – 6,259,176 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.75 |
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -19.70 |
| Energy contribution | -19.95 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6259067 109 - 22224390 ACGCGUAUCAAGUUACAACUUGCAACAAAUAUUAAAUCAGUAAAAUGUCCUUUAGGGCGCACUCCACUGUAAUUGUUGUUGCAGCCACCCGUAUUCCUUCCUUUUUUUC----------- ..(((((......)))....((((((((..((((...((((....(((((....)))))......))))))))..))))))))))........................----------- ( -20.60) >DroSec_CAF1 720 107 - 1 ACGCGUAUCAAGUUACAACUUGCAACAAAUAUUAAAUCAGUAAAAUGUCCUUUAGGGCGCACUCCACUGUAAUUGUUGUUGCAGCCACCCGUACUCCCUUCUU--UUCC----------- ..(((((......)))....((((((((..((((...((((....(((((....)))))......))))))))..))))))))))..................--....----------- ( -20.60) >DroSim_CAF1 728 107 - 1 ACGCGUAUCAAGUUACAAUUUGCAACAAAUAUUAAAUCAGUAAAAUGUCCUUUAGGGCGCACUCCACUGUAAUUGUUGUUGCAGCCACCCAUACUCCUUCCUU--UUCC----------- ..(((((......)))....((((((((..((((...((((....(((((....)))))......))))))))..))))))))))..................--....----------- ( -20.60) >DroYak_CAF1 238 118 - 1 ACGCGUAUCAAGUUACAACUUGCAACAAAUAUUAAAUCAGUAAAAUGUCCUUUAGGGCGCACUCCACUAUAAUUGUUGUUGCAGCCACCCGUACUCCUUCCUU--UUCUCCGCCUCUCCC ..(((((......)))...(((((((((..((((...........(((((....)))))..........))))..)))))))))...................--......))....... ( -17.90) >consensus ACGCGUAUCAAGUUACAACUUGCAACAAAUAUUAAAUCAGUAAAAUGUCCUUUAGGGCGCACUCCACUGUAAUUGUUGUUGCAGCCACCCGUACUCCUUCCUU__UUCC___________ ..(((((......)))....((((((((..((((...((((....(((((....)))))......))))))))..))))))))))................................... (-19.70 = -19.95 + 0.25)


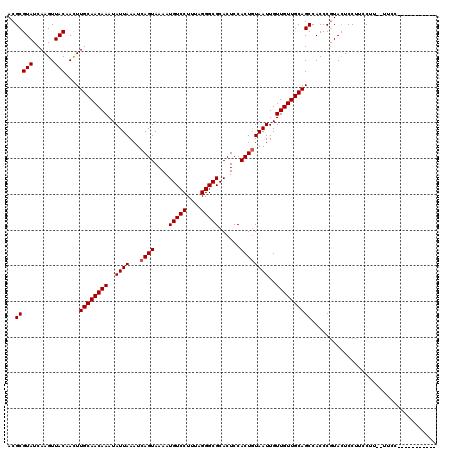
| Location | 6,259,096 – 6,259,212 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.79 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -33.19 |
| Energy contribution | -33.00 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6259096 116 + 22224390 AACAACAAUUACAGUGGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUACGCGUCGAGUUGCACGUUGCAAGGAGCAGC----GCUGUGGGACG ...........((((((((((..((.....)).))))))))))...........(((.((...(((((((((((....)))))))))))((((((.....)))))----)...)).))). ( -36.20) >DroSec_CAF1 747 120 + 1 AACAACAAUUACAGUGGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUACGCGUCAAGUUGCACGUCGCAAGGAGGGACGCAGGAUGUGGGACG ...........((((((((((..((.....)).))))))))))...........(((.((.(((((((((((((....)))))))))).((((.(.....).))))...))).)).))). ( -34.70) >DroSim_CAF1 755 116 + 1 AACAACAAUUACAGUGGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAAUUGUAACUUGAUACGCGUCGAGUUGCACGUCGCAAGAAGCAGC----GUUGUGGGACG .(((((.....((((((((((..((.....)).))))))))))...........(((((....(((((((((((....)))))))))))..((....)).)))))----)))))...... ( -39.60) >DroYak_CAF1 276 116 + 1 AACAACAAUUAUAGUGGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUACGCGUCGAGUUGCACGUCGCAACGAGCAGC----GCUGUGGGACG ..((((((.((((((((((((..((.....)).)))))))))......))).)))))).....(((((((((((....)))))))))))((((.((...(((...----))).)).)))) ( -33.50) >consensus AACAACAAUUACAGUGGAGUGCGCCCUAAAGGACAUUUUACUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUACGCGUCGAGUUGCACGUCGCAAGGAGCAGC____GCUGUGGGACG ..((((((.((((((((((((..((.....)).))))))))))......)).)))))).....(((((((((((....)))))))))))((((.((...(((.......))).)).)))) (-33.19 = -33.00 + -0.19)

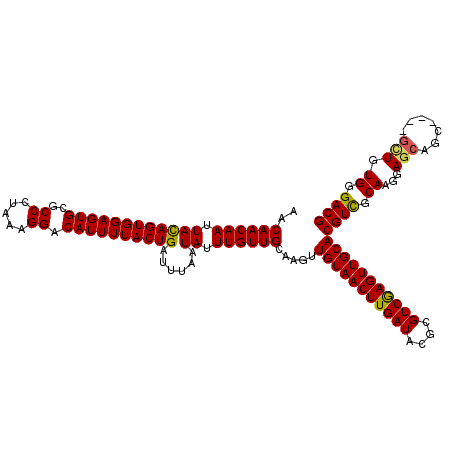
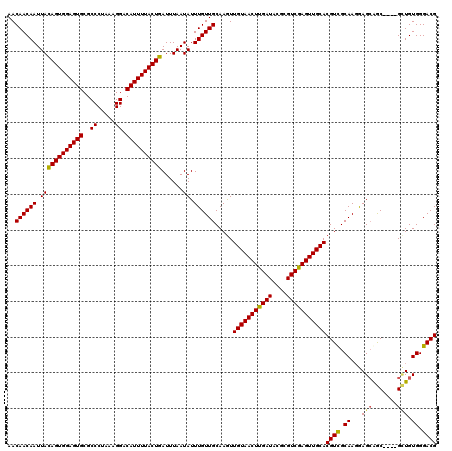
| Location | 6,259,136 – 6,259,252 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.79 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -37.88 |
| Energy contribution | -37.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.776685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6259136 116 + 22224390 CUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUACGCGUCGAGUUGCACGUUGCAAGGAGCAGC----GCUGUGGGACGCAGGAUGAGCCCAAGGAUCUGCCGGCUGCUUCUUGUGAGA .....................((.((((((((((....)))))))))))).(..(((((((((((----....((((...(.....)..)))).((.....)).)))))))))))..).. ( -41.00) >DroSec_CAF1 787 120 + 1 CUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUACGCGUCAAGUUGCACGUCGCAAGGAGGGACGCAGGAUGUGGGACGCAGGAUGAGCCCAAGGAUAUGCCGGCUGCUUCUUGUGAGA ....((..(((((..((((..((.((((((((((....))))))))))))(((.(.....).))))))))))))..))(((((((.(((((...((.....)))))).).)))))))... ( -37.00) >DroSim_CAF1 795 116 + 1 CUGAUUUAAUAUUUGUUGCAAAUUGUAACUUGAUACGCGUCGAGUUGCACGUCGCAAGAAGCAGC----GUUGUGGGACGCAGGAUGAGCCCAAGGAUCUGCCGGCUGCUUCUUGUGAGA .......................(((((((((((....)))))))))))..((((((((((((((----....((((...(.....)..)))).((.....)).)))))))))))))).. ( -43.80) >DroYak_CAF1 316 116 + 1 CUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUACGCGUCGAGUUGCACGUCGCAACGAGCAGC----GCUGUGGGACGCAGGAUGAGCCCAAGGAUCUGCCGGCUGCUUCUUGUGAGA .....................((.((((((((((....)))))))))))).((((((.(((((((----....((((...(.....)..)))).((.....)).))))))).)))))).. ( -38.10) >consensus CUGAUUUAAUAUUUGUUGCAAGUUGUAACUUGAUACGCGUCGAGUUGCACGUCGCAAGGAGCAGC____GCUGUGGGACGCAGGAUGAGCCCAAGGAUCUGCCGGCUGCUUCUUGUGAGA .......................(((((((((((....)))))))))))..((((((((((((((........((((...(.....)..)))).((.....)).)))))))))))))).. (-37.88 = -37.88 + 0.00)

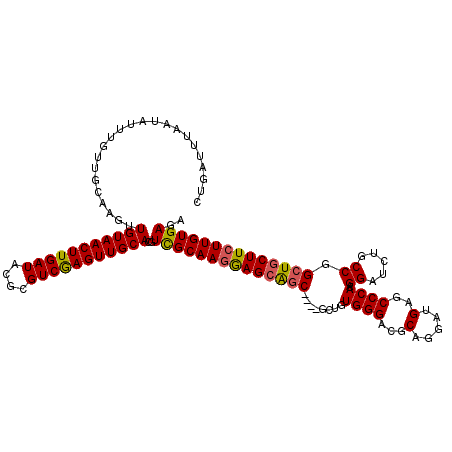
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:40 2006