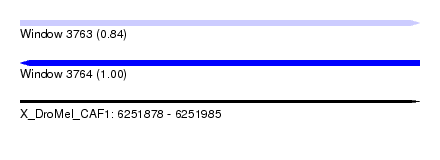
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,251,878 – 6,251,985 |
| Length | 107 |
| Max. P | 0.999793 |

| Location | 6,251,878 – 6,251,985 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 72.38 |
| Mean single sequence MFE | -37.39 |
| Consensus MFE | -20.66 |
| Energy contribution | -24.89 |
| Covariance contribution | 4.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6251878 107 + 22224390 UUGGUAUUGGUGCUGGUGUGCGGCUGCCAAAAUUCGAAUGCCCCGCCGUGCUCCGAAAGGCCGGGAAUGGGGCAUCCAUUUUCUUGUCCAACGCUUCCUGGUCAGUU ......((((.(((((((.(.(((...............))))))))).)).))))..(((((((((..(((((..........))))).....))))))))).... ( -37.96) >DroSec_CAF1 18578 100 + 1 UUGGU------GUUGGUGUGCGGCUGCCAAAAUUCGGAUGCCCC-CGCUGCUCCGAAAGUCCGGGAAAGGGGCAUCCAUUUUCUUGGCCGCCGCUUCCUGGCCAGUU (((((------...((...(((((.(((((.....(((((((((-.....(.(((......))))...)))))))))......))))).)))))..))..))))).. ( -48.00) >DroYak_CAF1 25762 83 + 1 UUGGU------GGCG--GUGCGGCUGCCAAAAUUCGGAUUCCCUCC---GCUCCAAAAGGCC------------UCCA-UUUUUAAGCCGCUGCUACCUGGUCAGUU ..(((------((((--..((((((...(((((..(((.....)))---(((......))).------------...)-))))..)))))))))))))......... ( -26.20) >consensus UUGGU______GCUGGUGUGCGGCUGCCAAAAUUCGGAUGCCCC_C__UGCUCCGAAAGGCCGGGAA_GGGGCAUCCAUUUUCUUGGCCGCCGCUUCCUGGUCAGUU ...........(((((...(((((.(((((.....(((((((((........((....))........)))))))))......))))).)))))...)))))..... (-20.66 = -24.89 + 4.23)



| Location | 6,251,878 – 6,251,985 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 72.38 |
| Mean single sequence MFE | -36.63 |
| Consensus MFE | -23.77 |
| Energy contribution | -26.56 |
| Covariance contribution | 2.79 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.65 |
| SVM decision value | 4.09 |
| SVM RNA-class probability | 0.999793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6251878 107 - 22224390 AACUGACCAGGAAGCGUUGGACAAGAAAAUGGAUGCCCCAUUCCCGGCCUUUCGGAGCACGGCGGGGCAUUCGAAUUUUGGCAGCCGCACACCAGCACCAAUACCAA ...............(((((.(((((...((((((((((....((((((.....).)).))).))))))))))..)))))((....))...)))))........... ( -33.20) >DroSec_CAF1 18578 100 - 1 AACUGGCCAGGAAGCGGCGGCCAAGAAAAUGGAUGCCCCUUUCCCGGACUUUCGGAGCAGCG-GGGGCAUCCGAAUUUUGGCAGCCGCACACCAAC------ACCAA ...(((...((..(((((.(((((((...(((((((((((..(((((....)))).)....)-))))))))))..))))))).)))))...))...------.))). ( -47.50) >DroYak_CAF1 25762 83 - 1 AACUGACCAGGUAGCAGCGGCUUAAAAA-UGGA------------GGCCUUUUGGAGC---GGAGGGAAUCCGAAUUUUGGCAGCCGCAC--CGCC------ACCAA .........(((.((.((((((..((((-((((------------..(((((((...)---))))))..)))..)))))...))))))..--.)).------))).. ( -29.20) >consensus AACUGACCAGGAAGCGGCGGCCAAGAAAAUGGAUGCCCC_UUCCCGGCCUUUCGGAGCA__G_GGGGCAUCCGAAUUUUGGCAGCCGCACACCAAC______ACCAA .............(((((.(((((((...((((((((((........((....))........))))))))))..))))))).)))))................... (-23.77 = -26.56 + 2.79)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:32 2006