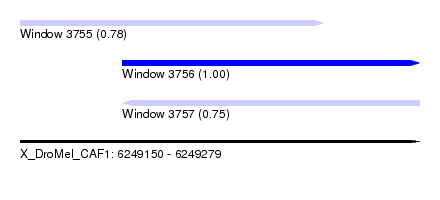
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,249,150 – 6,249,279 |
| Length | 129 |
| Max. P | 0.996578 |

| Location | 6,249,150 – 6,249,248 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.13 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -13.31 |
| Energy contribution | -13.20 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6249150 98 + 22224390 AUU-------GCCACAUGAAAAUUGCACACGGAAUGGACUCAUUAAACUUGCAACAUUUUGCGACUUUCGCAAUCAAUUUAUAAAAUAAUGUGCGC--AAGUACUUU------------- .((-------(((((((.....(((((.....((((....)))).....)))))....(((((.....)))))...............))))).))--)).......------------- ( -21.20) >DroSec_CAF1 15941 94 + 1 A-----------CCCAUGGAAACUGCACACGAAAUGGGCUCAUUAAACUUGCAACAUUUUGCGA--UUCGCAAUCAAUUUAUAUAAUAAAGUGCGCACAAGUACUUU------------- .-----------((((((....)..........)))))..........((((((....))))))--.....................(((((((......)))))))------------- ( -15.10) >DroYak_CAF1 23052 113 + 1 CCACUUCUGCGGCCCAUGAAUACUGCAGCGUGAAUGGGCUCAUUAAACUUGCAACAGU-UGCGA--UUCGCAAUCAAUUCAUAU--UAAUAUGCGC--AAGUACUUUAUGCUCGGCCACA ......((((((..........)))))).(((..(((((.......((((((....((-((((.--..)))))).....((((.--...)))).))--)))).......)))))..))). ( -29.64) >consensus A_________G_CCCAUGAAAACUGCACACGGAAUGGGCUCAUUAAACUUGCAACAUUUUGCGA__UUCGCAAUCAAUUUAUAUAAUAAUGUGCGC__AAGUACUUU_____________ ...............(((((...((((.....((((....)))).....))))......((((.....)))).....)))))........((((......))))................ (-13.31 = -13.20 + -0.11)

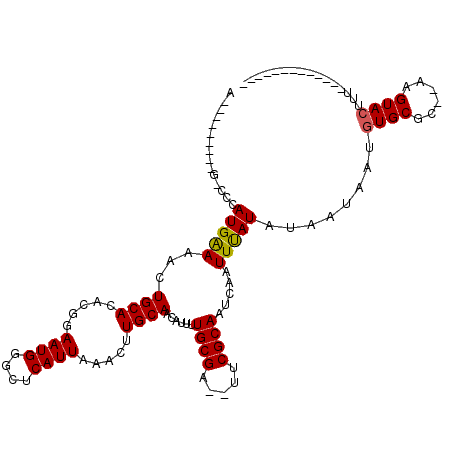

| Location | 6,249,183 – 6,249,279 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -15.20 |
| Energy contribution | -15.95 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6249183 96 + 22224390 CAUUAAACUUGCAACAUUUUGCGACUUUCGCAAUCAAUUUAUAAAAUAAUGUGCGC--AAGUACUUU-------------------AGCUGCAUUUUUAAUGACGAACUA---UUAGUGA (((((((..((((.(...(((((.....))))).............(((.((((..--..)))).))-------------------)).))))..)))))))........---....... ( -23.30) >DroSec_CAF1 15970 99 + 1 CAUUAAACUUGCAACAUUUUGCGA--UUCGCAAUCAAUUUAUAUAAUAAAGUGCGCACAAGUACUUU-------------------AGCUGCAUUUUUAAUGGCGCACUAUUAUUAGUGA (((((((..((((.(...(((((.--..))))).............((((((((......)))))))-------------------)).))))..)))))))...(((((....))))). ( -27.10) >DroEre_CAF1 10676 110 + 1 CAUUAAACUUGCAACAGU-UGCCA--CUCGCAAUCAAUUCAUAU--UAAUAUGCGC--AAGUACUUAAUACUCGGCCACAAUAAACAGCUGCAUUUUUAAUGGCAAACUA---UUAGAGA .(((((((((((....((-(((..--...))))).....((((.--...)))).))--))))..)))))...((((...........))))..((((((((((....)))---))))))) ( -19.00) >DroYak_CAF1 23092 110 + 1 CAUUAAACUUGCAACAGU-UGCGA--UUCGCAAUCAAUUCAUAU--UAAUAUGCGC--AAGUACUUUAUGCUCGGCCACAAUGAAGAGCUGCAGUUUUAAUGCCAAACUA---UUAGAGA (((((((.(((((...((-((((.--..))))))..........--(((..(((..--..)))..))).((((..(......)..))))))))).)))))))........---....... ( -23.10) >consensus CAUUAAACUUGCAACAGU_UGCGA__UUCGCAAUCAAUUCAUAU__UAAUAUGCGC__AAGUACUUU___________________AGCUGCAUUUUUAAUGGCAAACUA___UUAGAGA (((((((..((((.(....((((.....))))..................((((......)))).......................).))))..))))))).................. (-15.20 = -15.95 + 0.75)



| Location | 6,249,183 – 6,249,279 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -15.75 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
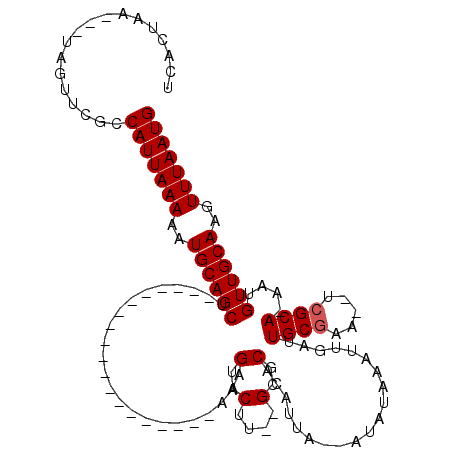
>X_DroMel_CAF1 6249183 96 - 22224390 UCACUAA---UAGUUCGUCAUUAAAAAUGCAGCU-------------------AAAGUACUU--GCGCACAUUAUUUUAUAAAUUGAUUGCGAAAGUCGCAAAAUGUUGCAAGUUUAAUG .......---........(((((((..((((((.-------------------...((((((--.((((.((((..........)))))))).)))).)).....))))))..))))))) ( -21.10) >DroSec_CAF1 15970 99 - 1 UCACUAAUAAUAGUGCGCCAUUAAAAAUGCAGCU-------------------AAAGUACUUGUGCGCACUUUAUUAUAUAAAUUGAUUGCGAA--UCGCAAAAUGUUGCAAGUUUAAUG .(((((....)))))...(((((((..(((((((-------------------(((((.(......).)))))).............(((((..--.)))))...))))))..))))))) ( -22.40) >DroEre_CAF1 10676 110 - 1 UCUCUAA---UAGUUUGCCAUUAAAAAUGCAGCUGUUUAUUGUGGCCGAGUAUUAAGUACUU--GCGCAUAUUA--AUAUGAAUUGAUUGCGAG--UGGCA-ACUGUUGCAAGUUUAAUG .......---........(((((((..((((((.((....(((.(((((((((...))))))--)((((.((((--((....)))))))))).)--).)))-)).))))))..))))))) ( -28.60) >DroYak_CAF1 23092 110 - 1 UCUCUAA---UAGUUUGGCAUUAAAACUGCAGCUCUUCAUUGUGGCCGAGCAUAAAGUACUU--GCGCAUAUUA--AUAUGAAUUGAUUGCGAA--UCGCA-ACUGUUGCAAGUUUAAUG .......---........(((((((..((((((......((((((....(((.........)--))(((.((((--((....)))))))))...--)))))-)..))))))..))))))) ( -23.50) >consensus UCACUAA___UAGUUCGCCAUUAAAAAUGCAGCU___________________AAAGUACUU__GCGCACAUUA__AUAUAAAUUGAUUGCGAA__UCGCA_AAUGUUGCAAGUUUAAUG ..................(((((((..((((((.......................(..(....)..)....................((((.....))))....))))))..))))))) (-15.75 = -16.00 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:26 2006