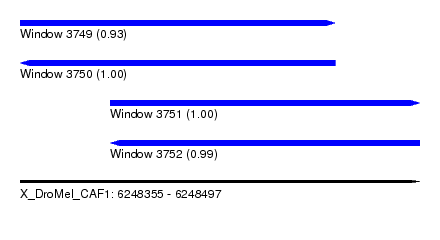
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,248,355 – 6,248,497 |
| Length | 142 |
| Max. P | 0.997404 |

| Location | 6,248,355 – 6,248,467 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.46 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -29.15 |
| Energy contribution | -28.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6248355 112 + 22224390 UGGGUGUGUAGGAUGUGUAG--------GAUGUGUAGGUGUGUGGAUGUGGGCAACGAUGGAGCUGAAAGGCUUAGCAGUUUGCUGAGCACACGCUCCACGGGGAAAUCUCCUUUGUUUU (((((((((...........--------......(((.(.(((.(.((....)).).))).).)))....(((((((.....))))))))))))).))).((((....))))........ ( -32.00) >DroEre_CAF1 9839 91 + 1 ----------------------------GGUGUGUGGG-AUGUGGAUGUGGGCAGCGAUGGAGCUGAAAGGCUUAGCAGUUUGCUGAGCACACGCUCCGCGGGGAAAUCUCCUGUUUUUU ----------------------------.....((((.-.((((........((((......))))....(((((((.....))))))).))))..))))((((....))))........ ( -34.50) >DroYak_CAF1 22256 113 + 1 UAGGUGUGU-----GUGCAGGAGUAUAGGAUGUGUAGG-AUGUGGAUGUGGGCAACGAUGGAGCUGAAAGGCUUAGCAGUUUGCUGAGCUCACGCUCCGCGGGGAAAUCUCCUUUGUUU- ......(.(-----((((....))))).).........-..((((((((((((.......(((((....)))))(((.....)))..))))))).)))))((((....)))).......- ( -35.30) >consensus U_GGUGUGU_____GUG_AG________GAUGUGUAGG_AUGUGGAUGUGGGCAACGAUGGAGCUGAAAGGCUUAGCAGUUUGCUGAGCACACGCUCCGCGGGGAAAUCUCCUUUGUUUU .........................................((((((((((((.........))).....(((((((.....))))))).)))).)))))((((....))))........ (-29.15 = -28.93 + -0.22)


| Location | 6,248,355 – 6,248,467 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.46 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -18.33 |
| Energy contribution | -18.33 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6248355 112 - 22224390 AAAACAAAGGAGAUUUCCCCGUGGAGCGUGUGCUCAGCAAACUGCUAAGCCUUUCAGCUCCAUCGUUGCCCACAUCCACACACCUACACAUC--------CUACACAUCCUACACACCCA ........(((........((((((((.((.(((.(((.....))).)))....)))))))).)).........)))...............--------.................... ( -21.83) >DroEre_CAF1 9839 91 - 1 AAAAAACAGGAGAUUUCCCCGCGGAGCGUGUGCUCAGCAAACUGCUAAGCCUUUCAGCUCCAUCGCUGCCCACAUCCACAU-CCCACACACC---------------------------- ........(((........((.(((((.((.(((.(((.....))).)))....)))))))..)).........)))....-..........---------------------------- ( -19.13) >DroYak_CAF1 22256 113 - 1 -AAACAAAGGAGAUUUCCCCGCGGAGCGUGAGCUCAGCAAACUGCUAAGCCUUUCAGCUCCAUCGUUGCCCACAUCCACAU-CCUACACAUCCUAUACUCCUGCAC-----ACACACCUA -......(((((..........(((((.((((((.(((.....))).)))...))))))))...((.....))........-...............)))))....-----......... ( -23.90) >consensus AAAACAAAGGAGAUUUCCCCGCGGAGCGUGUGCUCAGCAAACUGCUAAGCCUUUCAGCUCCAUCGUUGCCCACAUCCACAU_CCUACACAUC________CU_CAC_____ACACACC_A ........(((........((.(((((..(.(((.(((.....))).)))).....)))))..)).........)))........................................... (-18.33 = -18.33 + 0.00)



| Location | 6,248,387 – 6,248,497 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -45.58 |
| Consensus MFE | -41.79 |
| Energy contribution | -41.60 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6248387 110 + 22224390 UGUGGAUGUGGGCAACGAUGGAGCUGAAAGGCUUAGCAGUUUGCUGAGCACACGCUCCACGGGGAAAUCUCCUUUGUUUU----------UUUCCACCUCGCGCCCACUUUUUACGCGCA .(((((.((((((..(((((((((((....(((((((.....))))))).)).)))))).(((((((.............----------))))).))))).))))))..)))))..... ( -44.52) >DroSec_CAF1 15199 110 + 1 UGUGCAUGUGGGCAACGAUGGAGCUGAAAGGCUUAGCAGUUUGCUGAGCACACGCUCCACGGGGAAAUCUCCUUUGUUUU----------UUUCCACUUCGCGCCCACUUUUUACGCGCA .((((..((((((..((.((((((((....(((((((.....))))))).)).))))))))((((((.(......)...)----------))))).......)))))).......)))). ( -44.30) >DroEre_CAF1 9850 120 + 1 UGUGGAUGUGGGCAGCGAUGGAGCUGAAAGGCUUAGCAGUUUGCUGAGCACACGCUCCGCGGGGAAAUCUCCUGUUUUUUUUUUUUUUUGUUUCCACCUCGCGCCCACUUUUUACGCGCA .(((((.((((((.((((.(((((((....(((((((.....))))))).)).)))))..((((((((.....................)))))).))))))))))))..)))))..... ( -47.90) >DroYak_CAF1 22290 109 + 1 UGUGGAUGUGGGCAACGAUGGAGCUGAAAGGCUUAGCAGUUUGCUGAGCUCACGCUCCGCGGGGAAAUCUCCUUUGUUU-----------UUUCCACCUUGCGCCCACUUUUUGCGAGCA .(..((.((((((......((((((((...(((((((.....)))))))))).)))))(((((((((.(......)...-----------))))).))..))))))))..))..)..... ( -45.60) >consensus UGUGGAUGUGGGCAACGAUGGAGCUGAAAGGCUUAGCAGUUUGCUGAGCACACGCUCCACGGGGAAAUCUCCUUUGUUUU__________UUUCCACCUCGCGCCCACUUUUUACGCGCA .(((((.((((((..((.((((((((....(((((((.....))))))).)).))))))))((((....)))).............................))))))..)))))..... (-41.79 = -41.60 + -0.19)



| Location | 6,248,387 – 6,248,497 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -39.01 |
| Consensus MFE | -35.64 |
| Energy contribution | -35.95 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6248387 110 - 22224390 UGCGCGUAAAAAGUGGGCGCGAGGUGGAAA----------AAAACAAAGGAGAUUUCCCCGUGGAGCGUGUGCUCAGCAAACUGCUAAGCCUUUCAGCUCCAUCGUUGCCCACAUCCACA ............((((((((((((.(((((----------.............))))))).((((((.((.(((.(((.....))).)))....))))))))))).)))))))....... ( -39.72) >DroSec_CAF1 15199 110 - 1 UGCGCGUAAAAAGUGGGCGCGAAGUGGAAA----------AAAACAAAGGAGAUUUCCCCGUGGAGCGUGUGCUCAGCAAACUGCUAAGCCUUUCAGCUCCAUCGUUGCCCACAUGCACA .(.((((.....((((((((((.(.(((((----------.............))))).).((((((.((.(((.(((.....))).)))....))))))))))).))))))))))).). ( -38.32) >DroEre_CAF1 9850 120 - 1 UGCGCGUAAAAAGUGGGCGCGAGGUGGAAACAAAAAAAAAAAAAAACAGGAGAUUUCCCCGCGGAGCGUGUGCUCAGCAAACUGCUAAGCCUUUCAGCUCCAUCGCUGCCCACAUCCACA ............((((((((((((.(((((.......................)))))))..(((((.((.(((.(((.....))).)))....))))))).)))).))))))....... ( -40.40) >DroYak_CAF1 22290 109 - 1 UGCUCGCAAAAAGUGGGCGCAAGGUGGAAA-----------AAACAAAGGAGAUUUCCCCGCGGAGCGUGAGCUCAGCAAACUGCUAAGCCUUUCAGCUCCAUCGUUGCCCACAUCCACA ............((((((((..((.(((((-----------............)))))))))(((((.((((((.(((.....))).)))...))))))))......))))))....... ( -37.60) >consensus UGCGCGUAAAAAGUGGGCGCGAGGUGGAAA__________AAAACAAAGGAGAUUUCCCCGCGGAGCGUGUGCUCAGCAAACUGCUAAGCCUUUCAGCUCCAUCGUUGCCCACAUCCACA ............((((((((((((.(((((.......................)))))))..(((((.((.(((.(((.....))).)))....))))))).)))).))))))....... (-35.64 = -35.95 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:22 2006