
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,237,765 – 6,237,917 |
| Length | 152 |
| Max. P | 0.999578 |

| Location | 6,237,765 – 6,237,877 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.38 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -18.47 |
| Energy contribution | -19.07 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6237765 112 + 22224390 UUUAACAAGCAGAAUAAUGCAGCAAGUGGAAUGCGGACCAAACGCCCACCCAC--------UAUCCACAUCCCAGCCACAAUCGGCCCACUAUUGGCAUCAACACGAUGCCACCGACACU ........(((......)))....(((((...(((.......)))........--------.............(((......))))))))..(((((((.....)))))))........ ( -27.50) >DroSec_CAF1 10739 111 + 1 UUUAACAAGCAGAAUAAUGCAGCAAGUGGAAUGCGGACCACCCGG-CACCCAC--------UAUCCACAUCCCGGCCACAAUCCACCCACUAUUGGCAUCAACACGAUGCCACCGACACU ........(((......))).....(((((.((.((.....((((-.......--------..........)))))).)).))))).......(((((((.....)))))))........ ( -27.33) >DroSim_CAF1 13591 111 + 1 UUUAACAAGCAGAAUAAUGCAGCAAGUGGAAUGCGGACCACCCGU-CACCCAC--------UAUCCACAUCCCAGCCACAAUCCACCCACUAUUGGCAUCAACACGAUGCCACCGACACU ........(((......)))....(((((...((((.....))))-...))))--------)...............................(((((((.....)))))))........ ( -25.20) >DroEre_CAF1 5484 103 + 1 UUUAACAAGCAGAAUAAUGCAGCAAGUGGAAUGCGGACCACCCAC----------------GAUCCACAUCUCAGCCAGCAUCC-CCCACUAUUGGCAUCAACACGAUGCCACCGACACU ........(((......)))....(((((.((((((.....))..----------------(((....))).......))))..-.)))))..(((((((.....)))))))........ ( -25.00) >DroYak_CAF1 18503 118 + 1 UUUAACAAGCAGAAUAAUGCAGCAAGUGGAAUGCGGACCACCCGC-CGCCUACUAUCCUUCUAUCCACAAGCCAGCCACAAUCC-CCCACUAUUGACAUCAACACGAUGCCACCGACACU ........(((......))).((..(((((..((((.....))))-.(....)..........)))))..))............-........((.((((.....)))).))........ ( -18.40) >consensus UUUAACAAGCAGAAUAAUGCAGCAAGUGGAAUGCGGACCACCCGC_CACCCAC________UAUCCACAUCCCAGCCACAAUCC_CCCACUAUUGGCAUCAACACGAUGCCACCGACACU ........(((......))).....((((.(((.(((..........................))).))).....))))..............(((((((.....)))))))........ (-18.47 = -19.07 + 0.60)

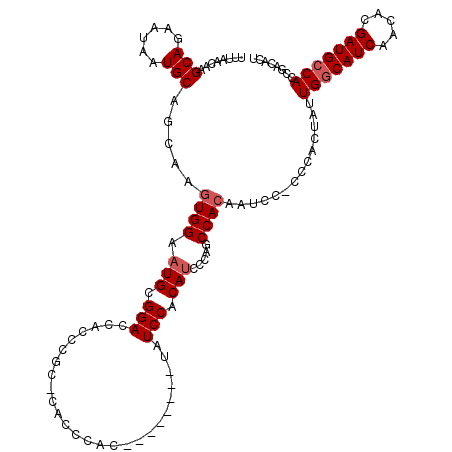

| Location | 6,237,765 – 6,237,877 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.38 |
| Mean single sequence MFE | -42.86 |
| Consensus MFE | -36.50 |
| Energy contribution | -38.06 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6237765 112 - 22224390 AGUGUCGGUGGCAUCGUGUUGAUGCCAAUAGUGGGCCGAUUGUGGCUGGGAUGUGGAUA--------GUGGGUGGGCGUUUGGUCCGCAUUCCACUUGCUGCAUUAUUCUGCUUGUUAAA ........(((((((.....))))))).(((..(((.((.((..((((((((((((((.--------..(.(....).)...))))))))))))...))..))....)).)))..))).. ( -44.20) >DroSec_CAF1 10739 111 - 1 AGUGUCGGUGGCAUCGUGUUGAUGCCAAUAGUGGGUGGAUUGUGGCCGGGAUGUGGAUA--------GUGGGUG-CCGGGUGGUCCGCAUUCCACUUGCUGCAUUAUUCUGCUUGUUAAA ........(((((((.....))))))).(((..((..((.((..((.((((((((((((--------.(.(...-.).).).)))))))))))....))..))....))..))..))).. ( -43.80) >DroSim_CAF1 13591 111 - 1 AGUGUCGGUGGCAUCGUGUUGAUGCCAAUAGUGGGUGGAUUGUGGCUGGGAUGUGGAUA--------GUGGGUG-ACGGGUGGUCCGCAUUCCACUUGCUGCAUUAUUCUGCUUGUUAAA ........(((((((.....))))))).(((..((..((.((..(((((((((((((((--------.(.(...-.).).).))))))))))))...))..))....))..))..))).. ( -48.00) >DroEre_CAF1 5484 103 - 1 AGUGUCGGUGGCAUCGUGUUGAUGCCAAUAGUGGG-GGAUGCUGGCUGAGAUGUGGAUC----------------GUGGGUGGUCCGCAUUCCACUUGCUGCAUUAUUCUGCUUGUUAAA ........(((((((.....)))))))..((..((-((((((.(((((..(((((((((----------------(....))))))))))..))...)))))))).)))..))....... ( -37.70) >DroYak_CAF1 18503 118 - 1 AGUGUCGGUGGCAUCGUGUUGAUGUCAAUAGUGGG-GGAUUGUGGCUGGCUUGUGGAUAGAAGGAUAGUAGGCG-GCGGGUGGUCCGCAUUCCACUUGCUGCAUUAUUCUGCUUGUUAAA ...((((((((((((.....)))))).(((((...-..))))).)))))).....(((((.((((((((..(((-((((((((........)))))))))))))))))))..)))))... ( -40.60) >consensus AGUGUCGGUGGCAUCGUGUUGAUGCCAAUAGUGGG_GGAUUGUGGCUGGGAUGUGGAUA________GUGGGUG_GCGGGUGGUCCGCAUUCCACUUGCUGCAUUAUUCUGCUUGUUAAA ........(((((((.....))))))).(((..((((((.((((((((((((((((((........................))))))))))))...))))))....))))))..))).. (-36.50 = -38.06 + 1.56)

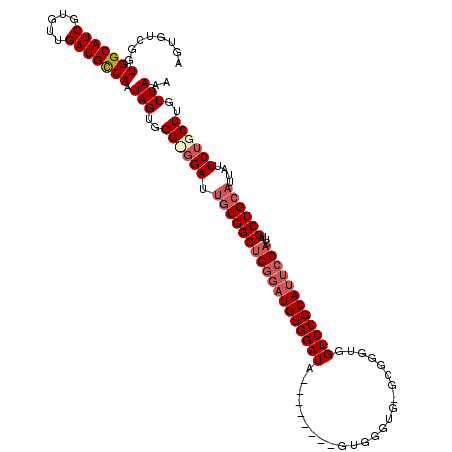
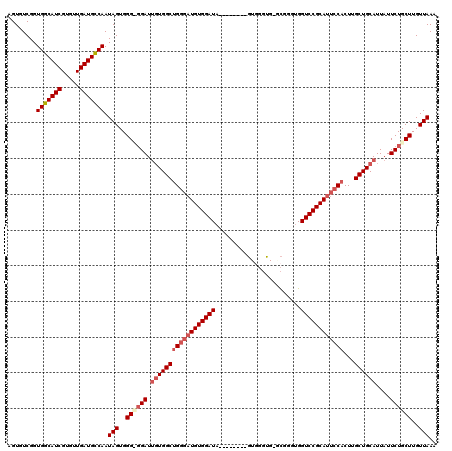
| Location | 6,237,805 – 6,237,917 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.03 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -19.10 |
| Energy contribution | -19.54 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6237805 112 + 22224390 AACGCCCACCCAC--------UAUCCACAUCCCAGCCACAAUCGGCCCACUAUUGGCAUCAACACGAUGCCACCGACACUUGCCUGUUCAUUGUUGUUUAUAAAAUUGGCAAGGAAUUUC ..((.........--------.............(((......))).......(((((((.....))))))).))...((((((.(((..((((.....))))))).))))))....... ( -23.90) >DroSec_CAF1 10779 111 + 1 CCCGG-CACCCAC--------UAUCCACAUCCCGGCCACAAUCCACCCACUAUUGGCAUCAACACGAUGCCACCGACACUUGCCUGUUCAUUGUUGUUUAUAAAAUUGGCAAGGAAUUUC .((((-.......--------..........))))......(((.........(((((((.....))))))).......(((((.(((..((((.....))))))).))))))))..... ( -23.43) >DroSim_CAF1 13631 111 + 1 CCCGU-CACCCAC--------UAUCCACAUCCCAGCCACAAUCCACCCACUAUUGGCAUCAACACGAUGCCACCGACACUUGCCUGUUCAUUGUUGUUUAUAAAAUUGGCAAGGAAUUUC .....-.......--------........(((..((((...............(((((((.....))))))).(((((..((......)).)))))..........))))..)))..... ( -22.30) >DroEre_CAF1 5524 103 + 1 CCCAC----------------GAUCCACAUCUCAGCCAGCAUCC-CCCACUAUUGGCAUCAACACGAUGCCACCGACACUUGCCUGUUCAUUGUUGUUUAUAAAAUUGGCAAGGAAUUUC .....----------------........(((..(((((.....-........(((((((.....))))))).(((((..((......)).))))).........)))))..)))..... ( -21.40) >DroYak_CAF1 18543 118 + 1 CCCGC-CGCCUACUAUCCUUCUAUCCACAAGCCAGCCACAAUCC-CCCACUAUUGACAUCAACACGAUGCCACCGACACUUGCCUGUUCAUUGUUGUUUAUAAAAUUGGCAAGGAAUUCC .....-.........(((((..........(((((.........-........((.((((.....)))).)).(((((..((......)).))))).........))))))))))..... ( -14.70) >consensus CCCGC_CACCCAC________UAUCCACAUCCCAGCCACAAUCC_CCCACUAUUGGCAUCAACACGAUGCCACCGACACUUGCCUGUUCAUUGUUGUUUAUAAAAUUGGCAAGGAAUUUC .............................(((..((((...............(((((((.....))))))).(((((..((......)).)))))..........))))..)))..... (-19.10 = -19.54 + 0.44)

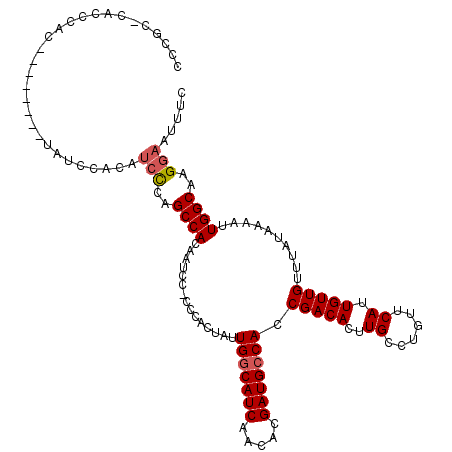
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:13 2006