
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,236,570 – 6,236,755 |
| Length | 185 |
| Max. P | 0.798200 |

| Location | 6,236,570 – 6,236,675 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.40 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -23.14 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6236570 105 - 22224390 AC--UGGGCCCACAGCUCAUUCCCGAAUCCGAUCAGGUUCGGUCCCAAAAGGAAAUCAAUUCAAAUUAGAUGAAAAUUAUAGUUGAAAUUGCCGGGGG-CG--------CCAUCCU---- ..--(((((((.(.((......(((((((......)))))))(((.....)))......(((((....(((....)))....)))))...)).).)))-).--------)))....---- ( -27.90) >DroSec_CAF1 9553 104 - 1 AC--UGGGCCCACGGCUCAUUCCCGAAUCCGAUCAUGUUCGGUCCCAAAAGGAUAUCAAUUCAAAUUAGAUGAAAAUUAUAGUUGAAAUUGCCGAGGG-CG--------CCCUC-U---- ..--.((((((.((((......((((((........))))))(((.....)))......(((((....(((....)))....)))))...)))).)))-).--------))...-.---- ( -29.60) >DroSim_CAF1 12433 104 - 1 AC--UGGGCCCACGGCUCAUUCCCGAAUCCGAUCAGGUUCGGUCCCAAAAGGAAAUCAAUUCAAAUUAGAUGAAAAUUAUAGUUGAAAUUGCCGAGGG-CG--------CCCUC-U---- ..--.((((((.((((......(((((((......)))))))(((.....)))......(((((....(((....)))....)))))...)))).)))-).--------))...-.---- ( -32.50) >DroEre_CAF1 4268 108 - 1 AC--UGGGCCCACAGCUCAUUCCCGAACCCGAUCAGGUUCGCUCCCAAAAGGAAAUCAAUUCAAAUUAGAUGAAAAUUAUAGUUGAAAUUGCCGGGGG-CG--------CCCUC-UGCUC ..--.((((...............(((((......)))))((((((....((....(((((..((((((((....))).)))))..))))))))))))-))--------)))..-..... ( -30.20) >DroYak_CAF1 17234 115 - 1 ACUCUGGGCACACAGCUCAUUCCCGAACCCGAUCAGGUUCGGUCCCAAAAGGAAAUCAAUUCAAAUUAGAUGAAAAUUAUAGUUGAAAUUGCCGGGGGGCGGAGAGGGCACCUC-U---- ....(((((.....))))).(((((((((......))))))(((((....((....(((((..((((((((....))).)))))..)))))))..))))))))((((...))))-.---- ( -35.40) >consensus AC__UGGGCCCACAGCUCAUUCCCGAAUCCGAUCAGGUUCGGUCCCAAAAGGAAAUCAAUUCAAAUUAGAUGAAAAUUAUAGUUGAAAUUGCCGGGGG_CG________CCCUC_U____ ....(((((.....)))))...(((((((......))))))).(((....((....(((((..((((((((....))).)))))..)))))))..)))...................... (-23.14 = -23.30 + 0.16)



| Location | 6,236,637 – 6,236,755 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.35 |
| Mean single sequence MFE | -38.24 |
| Consensus MFE | -31.90 |
| Energy contribution | -32.18 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6236637 118 + 22224390 GAACCUGAUCGGAUUCGGGAAUGAGCUGUGGGCCCA--GUAACUGUCUGUGAAUGGCUCAUUUAACGAGGUCAUGCGAUGACACUGGAAUCAUUUCUGGAUCCAUCCAGAGCGAUAAAGU ....(((((.((((((((((((((...(((((((..--...((.....))....)))))))....(.(((((((...))))).)).)..)))))))))))))))).)))........... ( -37.50) >DroSec_CAF1 9619 116 + 1 GAACAUGAUCGGAUUCGGGAAUGAGCCGUGGGCCCA--GUAACUGCCA--GAAUGGCUCAUUUAACGUGGUCAUGCGAUGACACUCGAAUCAUUUCUGGAUCCAUCCUGAGUGGAAAAGU ...((.((...((((((((((((((((((.(((...--......))).--..))))))))))....((.(((....))).)).))))))))...))))..(((((.....)))))..... ( -40.20) >DroSim_CAF1 12499 116 + 1 GAACCUGAUCGGAUUCGGGAAUGAGCCGUGGGCCCA--GUAACUGCCA--GAAUGGCUCAUUUAACGUGGUCAUGCGAUGACACUCGAAUCAUUUCUGGAUCCAUCCUGAGCGGAAAAGU ((.((.((...((((((((((((((((((.(((...--......))).--..))))))))))....((.(((....))).)).))))))))...)).)).))..(((.....)))..... ( -40.70) >DroEre_CAF1 4338 116 + 1 GAACCUGAUCGGGUUCGGGAAUGAGCUGUGGGCCCA--GUAACUGCCA--GAAUGGCUCAUUUAACGGGGUCAUGCGAUGACACUCGAAUCAUUUCUGGAUCCAUCCUCCUCGGAAAAGU ..........(((((((((((((((((((.(((...--......))).--..)))))))))))..(((((((((...))))).))))........)))))))).(((.....)))..... ( -39.10) >DroYak_CAF1 17309 105 + 1 GAACCUGAUCGGGUUCGGGAAUGAGCUGUGUGCCCAGAGUAACUGCCA--GAAUGGCUCAUUUGACGGGGUCAUGCGAUGACACUCGAAUCAUUUGUGGAUCCA-------------AGU ((((((....))))))(((((((((((((.....(((.....)))...--..)))))))))))..(((((((((...))))).))))..............)).-------------... ( -33.70) >consensus GAACCUGAUCGGAUUCGGGAAUGAGCUGUGGGCCCA__GUAACUGCCA__GAAUGGCUCAUUUAACGGGGUCAUGCGAUGACACUCGAAUCAUUUCUGGAUCCAUCCUGAGCGGAAAAGU ..........(((((((((((((((((((.(((...........))).....)))))))))))..(((((((((...))))).))))........))))))))................. (-31.90 = -32.18 + 0.28)

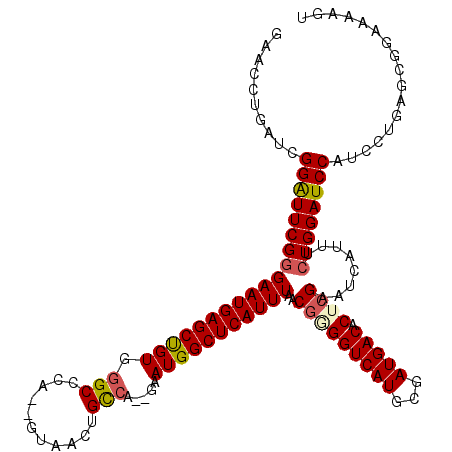
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:09 2006