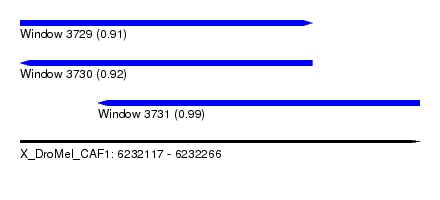
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,232,117 – 6,232,266 |
| Length | 149 |
| Max. P | 0.991579 |

| Location | 6,232,117 – 6,232,226 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.75 |
| Mean single sequence MFE | -36.17 |
| Consensus MFE | -22.52 |
| Energy contribution | -23.58 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6232117 109 + 22224390 ---------UAUGGUUCGAUG--GUGUAUGGUUGUGGAUAUGUGGUUAUAUCCAGCUUGCCUGCACCAAAUUAUUGCCUUUUACAAUUACAAUUGUAAUUGAGAAUGAGAGUUUUACGGC ---------..........((--(((((.(((.(((((((((....)))))))).)..)))))))))).......(((.....(((((((....)))))))(((((....)))))..))) ( -34.50) >DroSec_CAF1 5163 116 + 1 UG----GGGAAUGGUUCGAGGGGGCGUAGGGUUGUGGAUAUGUGGUUAUAUCCAGCUCGCCUGCACCAAAUUAUUGCCUUUUACAAUUACAAUUGUAAUUGAGAAUGAGAGUUUUACGAC .(----((.(((((((.(.(.(((((...(((..((((((((....)))))))))))))))).)..).))))))).)))....(((((((....)))))))(((((....)))))..... ( -34.60) >DroSim_CAF1 7235 116 + 1 UG----GGGAAUGGUUCGUGGGGGCGUGGGGUUGUGGAUAUGUGGUUAUAUCCAGCUCGCCUGCACCAAAUUAUUGCCUUUUACAAUUACAAUUGUAAUUGAGAAUGAGAGUUUUACGGC .(----((.(((((((.(((.(((((...(((..((((((((....)))))))))))))))).)))..))))))).)))....(((((((....)))))))(((((....)))))..... ( -39.20) >DroYak_CAF1 12605 110 + 1 GGGAAUGGGAAUGGUGCG--GUGGCGUAUGGUUGUGGAUU--------CAUGCAACUCGCCUGCGCCAAAUUAUUGCCUUUUACAAUUACAAUUGUAAUUGAGAAUGAGAGUUUUACGGC ((.(((((...(((((((--..((((...(((((((....--------..))))))))))))))))))..))))).)).....(((((((....)))))))(((((....)))))..... ( -36.40) >consensus UG____GGGAAUGGUUCGAGGGGGCGUAGGGUUGUGGAUAUGUGGUUAUAUCCAGCUCGCCUGCACCAAAUUAUUGCCUUUUACAAUUACAAUUGUAAUUGAGAAUGAGAGUUUUACGGC ............(((.......((((((.(((..((((((((....))))))))....)))))))))........))).....(((((((....)))))))................... (-22.52 = -23.58 + 1.06)

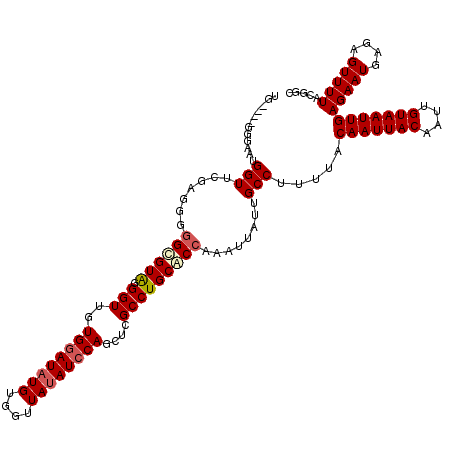
| Location | 6,232,117 – 6,232,226 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.75 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.18 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6232117 109 - 22224390 GCCGUAAAACUCUCAUUCUCAAUUACAAUUGUAAUUGUAAAAGGCAAUAAUUUGGUGCAGGCAAGCUGGAUAUAACCACAUAUCCACAACCAUACAC--CAUCGAACCAUA--------- ((((.....).....((..(((((((....)))))))..)).))).......(((((..((...(.(((((((......))))))))..))...)))--))..........--------- ( -22.30) >DroSec_CAF1 5163 116 - 1 GUCGUAAAACUCUCAUUCUCAAUUACAAUUGUAAUUGUAAAAGGCAAUAAUUUGGUGCAGGCGAGCUGGAUAUAACCACAUAUCCACAACCCUACGCCCCCUCGAACCAUUCCC----CA .(((...........((..(((((((....)))))))..))............((((.(((.(...(((((((......)))))))...)))).))))....))).........----.. ( -22.20) >DroSim_CAF1 7235 116 - 1 GCCGUAAAACUCUCAUUCUCAAUUACAAUUGUAAUUGUAAAAGGCAAUAAUUUGGUGCAGGCGAGCUGGAUAUAACCACAUAUCCACAACCCCACGCCCCCACGAACCAUUCCC----CA ((((.....).....((..(((((((....)))))))..)).)))......((.(((..((((.(.(((((((......)))))))......).))))..))).))........----.. ( -23.90) >DroYak_CAF1 12605 110 - 1 GCCGUAAAACUCUCAUUCUCAAUUACAAUUGUAAUUGUAAAAGGCAAUAAUUUGGCGCAGGCGAGUUGCAUG--------AAUCCACAACCAUACGCCAC--CGCACCAUUCCCAUUCCC ((((.....).....((..(((((((....)))))))..)).))).......(((.((.((((.((((..((--------....))))))....))))..--.)).)))........... ( -21.50) >consensus GCCGUAAAACUCUCAUUCUCAAUUACAAUUGUAAUUGUAAAAGGCAAUAAUUUGGUGCAGGCGAGCUGGAUAUAACCACAUAUCCACAACCAUACGCCCCCUCGAACCAUUCCC____CA ((((.....).....((..(((((((....)))))))..)).)))........((((..((...(.(((((((......))))))))..))...))))...................... (-16.92 = -17.18 + 0.25)



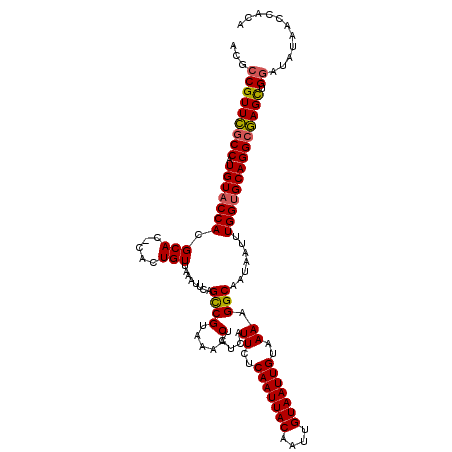
| Location | 6,232,146 – 6,232,266 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.05 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -27.42 |
| Energy contribution | -27.42 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6232146 120 - 22224390 ACGCCGUUUUCCAUGUACCACGCACCCACUGUUAAAUUCAGCCGUAAAACUCUCAUUCUCAAUUACAAUUGUAAUUGUAAAAGGCAAUAAUUUGGUGCAGGCAAGCUGGAUAUAACCACA ...((((((.((.(((((((.(((.....)))........((((.....).....((..(((((((....)))))))..)).))).......))))))))).)))).))........... ( -24.80) >DroSec_CAF1 5199 119 - 1 ACGCCGUUCGCCAUGUACCACGCAC-UACUGUUAAAUUCAGUCGUAAAACUCUCAUUCUCAAUUACAAUUGUAAUUGUAAAAGGCAAUAAUUUGGUGCAGGCGAGCUGGAUAUAACCACA ...(((((((((.(((((((...((-.((((.......)))).))..........((..(((((((....)))))))..))...........)))))))))))))).))........... ( -34.00) >DroSim_CAF1 7271 119 - 1 ACGCCGUUCGCCAUGUACCACGCAC-CACUGUUAAAUUCAGCCGUAAAACUCUCAUUCUCAAUUACAAUUGUAAUUGUAAAAGGCAAUAAUUUGGUGCAGGCGAGCUGGAUAUAACCACA ...(((((((((.(((((((.(((.-...)))........((((.....).....((..(((((((....)))))))..)).))).......)))))))))))))).))........... ( -33.20) >DroYak_CAF1 12643 111 - 1 ACGCCGUUCGCCAUGUACCACGCAC-CACUGUUAAACUCAGCCGUAAAACUCUCAUUCUCAAUUACAAUUGUAAUUGUAAAAGGCAAUAAUUUGGCGCAGGCGAGUUGCAUG-------- ..((..((((((.((..(((.(((.-...)))........((((.....).....((..(((((((....)))))))..)).))).......)))..))))))))..))...-------- ( -28.40) >consensus ACGCCGUUCGCCAUGUACCACGCAC_CACUGUUAAAUUCAGCCGUAAAACUCUCAUUCUCAAUUACAAUUGUAAUUGUAAAAGGCAAUAAUUUGGUGCAGGCGAGCUGGAUAUAACCACA ...(((((((((.(((((((.(((.....)))........((((.....).....((..(((((((....)))))))..)).))).......)))))))))))))).))........... (-27.42 = -27.42 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:02 2006