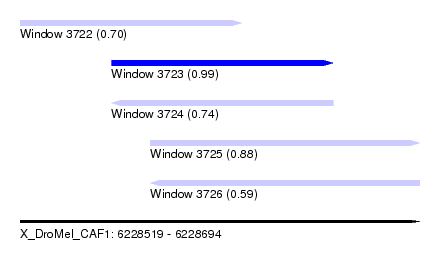
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,228,519 – 6,228,694 |
| Length | 175 |
| Max. P | 0.988436 |

| Location | 6,228,519 – 6,228,616 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 95.88 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -26.17 |
| Energy contribution | -26.40 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6228519 97 + 22224390 CGCGCAUAGUAAAUUGAAUUUUGUUGCCUUCUGCGGCGGCCAACAUCUCCUCCACUUUUGCCACCCACUUGUUUUGGUUUCUGUGGAGGCGACUCCA ......................((((((......)))))).....((.(((((((....((((...........))))....))))))).))..... ( -25.90) >DroSec_CAF1 2911 97 + 1 CGCGCAUAGUAAAUUGAAUUUUUUUGCCUUCUGCGGCGGCCAACAUCUCCUCUGCUUUUGCCACCAACUUGUUUUGGUUUCUGUGGAGGAGACUCCA ((((((..(((((.........)))))....))).))).......(((((((..(....((((...........))))....)..)))))))..... ( -26.80) >DroSim_CAF1 4975 97 + 1 CGCGCAUAGUAAAUUGAAUUUUGUUGCCUUCUGCGGCGGCCAACAUCUCCUCCGCUUUUGCCACCCACUUGGUUUGGUUUCUGUGGAGGAGACUCCA ......................((((((......)))))).....((((((((((....((((.((....))..))))....))))))))))..... ( -32.90) >consensus CGCGCAUAGUAAAUUGAAUUUUGUUGCCUUCUGCGGCGGCCAACAUCUCCUCCGCUUUUGCCACCCACUUGUUUUGGUUUCUGUGGAGGAGACUCCA ......................((((((......)))))).....((((((((((....((((...........))))....))))))))))..... (-26.17 = -26.40 + 0.23)


| Location | 6,228,559 – 6,228,656 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 95.88 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -24.39 |
| Energy contribution | -24.07 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.28 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6228559 97 + 22224390 CAACAUCUCCUCCACUUUUGCCACCCACUUGUUUUGGUUUCUGUGGAGGCGACUCCACUUUAUCACAUUUGUAUAAAUAUAUUACCCAAGUGUACUU .....((.(((((((....((((...........))))....))))))).))............(((((((..(((.....)))..))))))).... ( -24.10) >DroSec_CAF1 2951 97 + 1 CAACAUCUCCUCUGCUUUUGCCACCAACUUGUUUUGGUUUCUGUGGAGGAGACUCCACUUUAUCACAUUUGUAUAAAUAUAUUACCUAAGUGUACUU .....(((((((..(....((((...........))))....)..)))))))............(((((((..(((.....)))..))))))).... ( -22.70) >DroSim_CAF1 5015 97 + 1 CAACAUCUCCUCCGCUUUUGCCACCCACUUGGUUUGGUUUCUGUGGAGGAGACUCCACUUUAUCACAUUUGUAUAAAUAUAUUACCUAAGUGUACUU .....((((((((((....((((.((....))..))))....))))))))))............(((((((..(((.....)))..))))))).... ( -28.40) >consensus CAACAUCUCCUCCGCUUUUGCCACCCACUUGUUUUGGUUUCUGUGGAGGAGACUCCACUUUAUCACAUUUGUAUAAAUAUAUUACCUAAGUGUACUU .....((((((((((....((((...........))))....))))))))))............(((((((..(((.....)))..))))))).... (-24.39 = -24.07 + -0.33)


| Location | 6,228,559 – 6,228,656 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.88 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -20.19 |
| Energy contribution | -20.63 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.739156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6228559 97 - 22224390 AAGUACACUUGGGUAAUAUAUUUAUACAAAUGUGAUAAAGUGGAGUCGCCUCCACAGAAACCAAAACAAGUGGGUGGCAAAAGUGGAGGAGAUGUUG .....(((((......(((((((....)))))))...)))))..(((.(((((((.....(((...(....)..))).....))))))).))).... ( -24.00) >DroSec_CAF1 2951 97 - 1 AAGUACACUUAGGUAAUAUAUUUAUACAAAUGUGAUAAAGUGGAGUCUCCUCCACAGAAACCAAAACAAGUUGGUGGCAAAAGCAGAGGAGAUGUUG .....(((((...((.(((((((....))))))).)))))))..((((((((.......(((((......))))).((....)).)))))))).... ( -23.40) >DroSim_CAF1 5015 97 - 1 AAGUACACUUAGGUAAUAUAUUUAUACAAAUGUGAUAAAGUGGAGUCUCCUCCACAGAAACCAAACCAAGUGGGUGGCAAAAGCGGAGGAGAUGUUG .....(((((...((.(((((((....))))))).)))))))..(((((((((.(.....(((..((....)).))).....).))))))))).... ( -25.70) >consensus AAGUACACUUAGGUAAUAUAUUUAUACAAAUGUGAUAAAGUGGAGUCUCCUCCACAGAAACCAAAACAAGUGGGUGGCAAAAGCGGAGGAGAUGUUG .....(((((...((.(((((((....))))))).)))))))..(((((((((......(((..........))).((....))))))))))).... (-20.19 = -20.63 + 0.45)

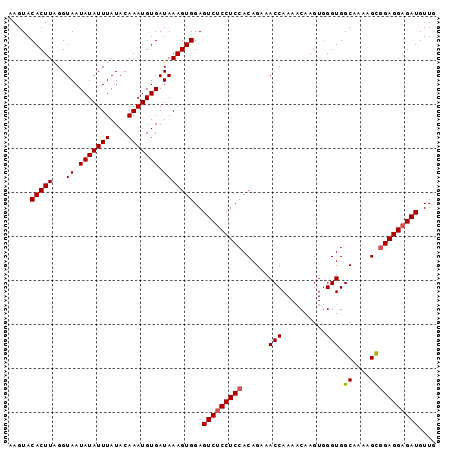
| Location | 6,228,576 – 6,228,694 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -21.97 |
| Energy contribution | -22.59 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6228576 118 + 22224390 UUGCCACCCACUUGUUUUGGUUUCUGUGGAGGCGACUCCACUUUAUCACAUUUGUAUAAAUAUAUUACCCAAGUGUACUUGAAGAAAAACCAGUGGUUUACGGAAAAUAGCUUUAU--UA .......(((((.(((((..((((.((((((....))))))......(((((((..(((.....)))..)))))))....)))).))))).)))))....................--.. ( -30.70) >DroSec_CAF1 2968 118 + 1 UUGCCACCAACUUGUUUUGGUUUCUGUGGAGGAGACUCCACUUUAUCACAUUUGUAUAAAUAUAUUACCUAAGUGUACUUGAAGAAAAACCAGUGCUUUACGGAAAAUGGUUUUAU--UA ..((((((.....((.(((((((((((((((....))))))......(((((((..(((.....)))..)))))))......)))..)))))).)).....))....)))).....--.. ( -28.10) >DroSim_CAF1 5032 118 + 1 UUGCCACCCACUUGGUUUGGUUUCUGUGGAGGAGACUCCACUUUAUCACAUUUGUAUAAAUAUAUUACCUAAGUGUACUUGAAGAAAAACCAGUGCUUUACGGAAAAUGGUUUUAU--UA ..((((((((((.(((((..((((.((((((....))))))......(((((((..(((.....)))..)))))))....))))..)))))))))......))....)))).....--.. ( -31.00) >DroYak_CAF1 10576 117 + 1 UUGCCACCCACUUGUUUUGGUUUGCGUGGAGGAGACUCCACUUUAUCACAUUUGUAUAAAUAUAUUACCCAAGUGCACUGAAAGAAAAAUAAGU---UCAAAAAGAUUGGUUUUUUCAUA ..((((...(((((((((..(((..((((((....)))))).......((((((..(((.....)))..)))))).....)))..)))))))))---((.....)).))))......... ( -26.60) >consensus UUGCCACCCACUUGUUUUGGUUUCUGUGGAGGAGACUCCACUUUAUCACAUUUGUAUAAAUAUAUUACCCAAGUGUACUUGAAGAAAAACCAGUGCUUUACGGAAAAUGGUUUUAU__UA ..((((((........(((((((..((((((....))))))......(((((((..(((.....)))..)))))))..........)))))))........))....))))......... (-21.97 = -22.59 + 0.62)

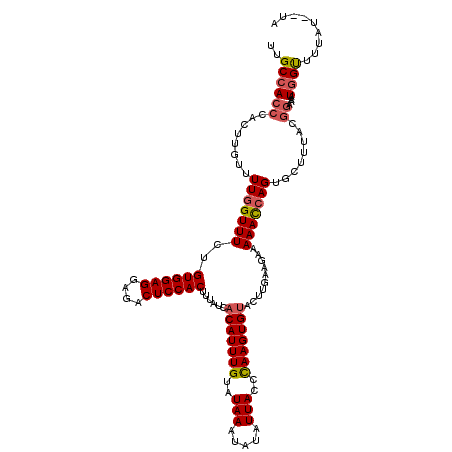

| Location | 6,228,576 – 6,228,694 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -19.40 |
| Energy contribution | -19.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6228576 118 - 22224390 UA--AUAAAGCUAUUUUCCGUAAACCACUGGUUUUUCUUCAAGUACACUUGGGUAAUAUAUUUAUACAAAUGUGAUAAAGUGGAGUCGCCUCCACAGAAACCAAAACAAGUGGGUGGCAA ..--.....(((((..........(((((.(((((....((((....))))(((..(((((((....))))))).....((((((....))))))....)))))))).)))))))))).. ( -30.10) >DroSec_CAF1 2968 118 - 1 UA--AUAAAACCAUUUUCCGUAAAGCACUGGUUUUUCUUCAAGUACACUUAGGUAAUAUAUUUAUACAAAUGUGAUAAAGUGGAGUCUCCUCCACAGAAACCAAAACAAGUUGGUGGCAA ..--......(((((..(.(.....)..((((((((.........(((....(((.((....)))))....))).....((((((....))))))))))))))......)..)))))... ( -22.30) >DroSim_CAF1 5032 118 - 1 UA--AUAAAACCAUUUUCCGUAAAGCACUGGUUUUUCUUCAAGUACACUUAGGUAAUAUAUUUAUACAAAUGUGAUAAAGUGGAGUCUCCUCCACAGAAACCAAACCAAGUGGGUGGCAA ..--......(((((....(.(((((....))))).)........(((((.(((..(((((((....))))))).....((((((....)))))).........)))))))))))))... ( -24.50) >DroYak_CAF1 10576 117 - 1 UAUGAAAAAACCAAUCUUUUUGA---ACUUAUUUUUCUUUCAGUGCACUUGGGUAAUAUAUUUAUACAAAUGUGAUAAAGUGGAGUCUCCUCCACGCAAACCAAAACAAGUGGGUGGCAA .........(((.......((((---(...........)))))..(((((((((..(((((((....))))))).....((((((....))))))....)))....)))))))))..... ( -22.70) >consensus UA__AUAAAACCAUUUUCCGUAAAGCACUGGUUUUUCUUCAAGUACACUUAGGUAAUAUAUUUAUACAAAUGUGAUAAAGUGGAGUCUCCUCCACAGAAACCAAAACAAGUGGGUGGCAA .........(((..............(((.(........).))).(((((((((..(((((((....))))))).....((((((....))))))....)))....)))))))))..... (-19.40 = -19.40 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:58 2006