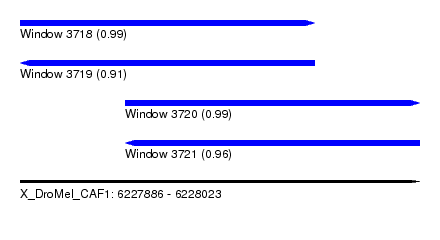
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,227,886 – 6,228,023 |
| Length | 137 |
| Max. P | 0.990573 |

| Location | 6,227,886 – 6,227,987 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.41 |
| Mean single sequence MFE | -39.58 |
| Consensus MFE | -24.74 |
| Energy contribution | -24.30 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6227886 101 + 22224390 UUUUCAAACAUUUUGCGCGUAAAGGUUUCC----UGUGGCGAAAUGACCUCUGA--CCGGGCGGAGUCAGAGGUCGUGGGGCUUACUGACUGAUGGCCGAAAAAUUA------------- (((((..(((((((((.((...(((...))----).)))))))))(((((((((--((.....).))))))))))))..((((...........)))))))))....------------- ( -33.10) >DroSec_CAF1 2274 116 + 1 UUUUCAACCAUUUUGCGCGUAAAGGUUUCC----UCUGGCGAAAUUACCUCUGAGGUCAGAGGGGGUCAGAGGUCGUGGGGCUUACUGACUGAUGGCCGAAAAAUUAGUACAGUAAGCCC ...............((((.........((----((((((.......((((((....))))))..)))))))).))))((((((((((((((((.........)))))).)))))))))) ( -46.40) >DroSim_CAF1 4338 116 + 1 UUUUCAACCAUUUUGCGCGUAAAGGUUUCC----UCUGGCGAAAUGACCUCUGAGGUCAGAGGGGGUCAGAGGUCGUGGGGCUUACUGACUGAUGGCCGAAAAAUUAGUACAGUAAGCCC ...............((((.........((----((((((.......((((((....))))))..)))))))).))))((((((((((((((((.........)))))).)))))))))) ( -46.40) >DroYak_CAF1 9912 102 + 1 UUUUCA-UCACCUUGC---UGAAGGUUUCCAUCAUCUGGU---AUGACCGC---------CAGGGGUCAGAGGUCGUGGGACUUACCGACUGAUGGCCGAAAAAUUAGUACAGUAAGC-- ......-....(((((---((..((((.(((.....))).---..))))((---------((..((((.((((((....))))).).))))..)))).............))))))).-- ( -32.40) >consensus UUUUCAACCAUUUUGCGCGUAAAGGUUUCC____UCUGGCGAAAUGACCUCUGA__UCAGAGGGGGUCAGAGGUCGUGGGGCUUACUGACUGAUGGCCGAAAAAUUAGUACAGUAAGC__ (((((....(((((.......)))))...........(((.......((((((....)))))).(((((((((((....))))).))))))....))))))))................. (-24.74 = -24.30 + -0.44)


| Location | 6,227,886 – 6,227,987 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.41 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -18.16 |
| Energy contribution | -20.10 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6227886 101 - 22224390 -------------UAAUUUUUCGGCCAUCAGUCAGUAAGCCCCACGACCUCUGACUCCGCCCGG--UCAGAGGUCAUUUCGCCACA----GGAAACCUUUACGCGCAAAAUGUUUGAAAA -------------....((((((..(((..((..(((((......(((((((((((......))--))))))))).((((......----))))...)))))..))...)))..)))))) ( -28.00) >DroSec_CAF1 2274 116 - 1 GGGCUUACUGUACUAAUUUUUCGGCCAUCAGUCAGUAAGCCCCACGACCUCUGACCCCCUCUGACCUCAGAGGUAAUUUCGCCAGA----GGAAACCUUUACGCGCAAAAUGGUUGAAAA ((((((((((.(((...............)))))))))))))..(((((........((((((....))))))...((((((.(((----((...)))))..))).)))..))))).... ( -38.96) >DroSim_CAF1 4338 116 - 1 GGGCUUACUGUACUAAUUUUUCGGCCAUCAGUCAGUAAGCCCCACGACCUCUGACCCCCUCUGACCUCAGAGGUCAUUUCGCCAGA----GGAAACCUUUACGCGCAAAAUGGUUGAAAA ((((((((((.(((...............)))))))))))))..(((((..(((((...((((....)))))))))((((((.(((----((...)))))..))).)))..))))).... ( -39.16) >DroYak_CAF1 9912 102 - 1 --GCUUACUGUACUAAUUUUUCGGCCAUCAGUCGGUAAGUCCCACGACCUCUGACCCCUG---------GCGGUCAU---ACCAGAUGAUGGAAACCUUCA---GCAAGGUGA-UGAAAA --...............((((((.((((((...((((.(((....)))...(((((....---------..))))))---)))...))))))..(((((..---..)))))..-)))))) ( -25.80) >consensus __GCUUACUGUACUAAUUUUUCGGCCAUCAGUCAGUAAGCCCCACGACCUCUGACCCCCUCUGA__UCAGAGGUCAUUUCGCCAGA____GGAAACCUUUACGCGCAAAAUGGUUGAAAA .................(((((((((((..((..((((((.....(((((((((............))))))))).....))........(....)..))))..))...))))))))))) (-18.16 = -20.10 + 1.94)

| Location | 6,227,922 – 6,228,023 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.79 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -21.76 |
| Energy contribution | -26.32 |
| Covariance contribution | 4.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6227922 101 + 22224390 GAAAUGACCUCUGA--CCGGGCGGAGUCAGAGGUCGUGGGGCUUACUGACUGAUGGCCGAAAAAUUA-----------------CUAAGACUGACAAUUAGAUAAUGCGAGACGGGCUUA .....(((((((((--((.....).))))))))))((.((.....)).))....(.(((....((((-----------------((((.........)))).))))......))).)... ( -28.00) >DroSec_CAF1 2310 120 + 1 GAAAUUACCUCUGAGGUCAGAGGGGGUCAGAGGUCGUGGGGCUUACUGACUGAUGGCCGAAAAAUUAGUACAGUAAGCCCCGCACUAAGACUGACAAUUAGAUAAUGCGAGGCGGGCUUA .......((((((....))))))..(((((.(((.(((((((((((((((((((.........)))))).))))))))))))))))....)))))...........((.......))... ( -48.90) >DroSim_CAF1 4374 120 + 1 GAAAUGACCUCUGAGGUCAGAGGGGGUCAGAGGUCGUGGGGCUUACUGACUGAUGGCCGAAAAAUUAGUACAGUAAGCCCCGCACUAAGACUGACAAUUAGAUAAUGCGAGGCGGGCUUA .....(((((((((..((....))..)))))))))(((((((((((((((((((.........)))))).)))))))))))))..((((.(((...(((....)))......))).)))) ( -49.30) >DroYak_CAF1 9948 96 + 1 ---AUGACCGC---------CAGGGGUCAGAGGUCGUGGGACUUACCGACUGAUGGCCGAAAAAUUAGUACAGUAAGC------------CUGAUAAUUAGAUAAUGCGAGGCUGGCUUA ---......((---------(((.(((((..(((((.(.......))))))..)))))....(((((...(((.....------------))).))))).............)))))... ( -27.00) >consensus GAAAUGACCUCUGA__UCAGAGGGGGUCAGAGGUCGUGGGGCUUACUGACUGAUGGCCGAAAAAUUAGUACAGUAAGC______CUAAGACUGACAAUUAGAUAAUGCGAGGCGGGCUUA .......((((((....))))))(((((.(...(((((((((((((((((((((.........)))))).))))))))))).........((((...)))).....))))..).))))). (-21.76 = -26.32 + 4.56)


| Location | 6,227,922 – 6,228,023 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.79 |
| Mean single sequence MFE | -32.01 |
| Consensus MFE | -16.16 |
| Energy contribution | -21.16 |
| Covariance contribution | 5.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6227922 101 - 22224390 UAAGCCCGUCUCGCAUUAUCUAAUUGUCAGUCUUAG-----------------UAAUUUUUCGGCCAUCAGUCAGUAAGCCCCACGACCUCUGACUCCGCCCGG--UCAGAGGUCAUUUC .......((...((.(((.((.((((...(((..((-----------------.....))..)))...)))).)))))))...))(((((((((((......))--)))))))))..... ( -25.90) >DroSec_CAF1 2310 120 - 1 UAAGCCCGCCUCGCAUUAUCUAAUUGUCAGUCUUAGUGCGGGGCUUACUGUACUAAUUUUUCGGCCAUCAGUCAGUAAGCCCCACGACCUCUGACCCCCUCUGACCUCAGAGGUAAUUUC ...((.......))...........(((((......((.(((((((((((.(((...............)))))))))))))).))....)))))..((((((....))))))....... ( -38.96) >DroSim_CAF1 4374 120 - 1 UAAGCCCGCCUCGCAUUAUCUAAUUGUCAGUCUUAGUGCGGGGCUUACUGUACUAAUUUUUCGGCCAUCAGUCAGUAAGCCCCACGACCUCUGACCCCCUCUGACCUCAGAGGUCAUUUC ............((((((.((.......))...))))))(((((((((((.(((...............))))))))))))))..(((((((((............)))))))))..... ( -42.06) >DroYak_CAF1 9948 96 - 1 UAAGCCAGCCUCGCAUUAUCUAAUUAUCAG------------GCUUACUGUACUAAUUUUUCGGCCAUCAGUCGGUAAGUCCCACGACCUCUGACCCCUG---------GCGGUCAU--- ...((((((((.................))------------)))..................((((...(((((...(((....)))..)))))...))---------)))))...--- ( -21.13) >consensus UAAGCCCGCCUCGCAUUAUCUAAUUGUCAGUCUUAG______GCUUACUGUACUAAUUUUUCGGCCAUCAGUCAGUAAGCCCCACGACCUCUGACCCCCUCUGA__UCAGAGGUCAUUUC ...((.......)).........................(((((((((((.(((...............))))))))))))))..(((((((((............)))))))))..... (-16.16 = -21.16 + 5.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:54 2006