| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,217,856 – 6,217,960 |
| Length | 104 |
| Max. P | 0.620956 |

| Location | 6,217,856 – 6,217,960 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.91 |
| Mean single sequence MFE | -36.09 |
| Consensus MFE | -23.44 |
| Energy contribution | -24.25 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
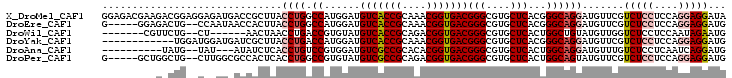
>X_DroMel_CAF1 6217856 104 + 22224390 GGAGACGAAGACGGAGGAGAUGACCGCUUACCUGGCCAUGGAUGUCACCGCAAACGGUGACGGGCGUGCUCACGGGCAGGAUGUUCGUCUCCUCCAGGAGGAUA (....)......(((((((((((..((...((((.((......(((((((....)))))))(((....)))..)).))))..)))))))))))))......... ( -48.30) >DroEre_CAF1 3005 97 + 1 G-----GGAGACUG--CCAAUAACCACUUACCUGGCCAUGGAUGUCACCGCAAACGGUGACGGGCGUGCUCACGGGCAGGAUGUUCGUCUCCUCCAGGAGGAUG .-----((((((((--........))....((((.((......(((((((....)))))))(((....)))..)).))))......))))))(((....))).. ( -35.70) >DroWil_CAF1 6630 89 + 1 -------CGUUCUG--CU------AACUAACCUGACCGUGUAUGUCACCGCAGACGGUGACGGGCGUGCUCACUGGCUGUAUGUUGGUCUCCUCCAAUAGAAUG -------(((((((--..------.........((((.....((((((((....))))))))(((((((.........)))))))))))........))))))) ( -27.11) >DroYak_CAF1 3145 92 + 1 ------------UGGAUGGAUGAUCGCUUACCUGACCAUGGAUGUCACCGCAAACGGUGACGGGCGUGCUCACGGGCAGGAUGUUCGUCUCCUCCAGGAGGAUG ------------..(((((((.(((.((..(((((.((((..((((((((....))))))))..)))).))).))..))))))))))))(((((...))))).. ( -37.40) >DroAna_CAF1 3801 89 + 1 ----------UAUG--UAU---AUAUCUCACCUGUCCGUGGAUGUCGCCGCACACGGUGACGGGCGUGCUCACUGGCAGGAUGUUUGUCUCCUCAAUCAGGAUG ----------....--...---........((((((.((....(((((((....)))))))(((....))))).)))))).........((((.....)))).. ( -27.10) >DroPer_CAF1 4102 97 + 1 G-----GCUGGCUG--CUUGGCGCCACUCACCUGGCCGUGUAUGUCGCCGCAGACGGUGACGGGCGUGCUCACUGGCAGUAUGUUCGUCUCCUCCAGGAGGAUG .-----((((.(((--(..((((.((..(((......)))..)).)))))))).))))(((((((((((((....).))))))))))))(((((...))))).. ( -40.90) >consensus _________GACUG__CAGAU_ACCACUUACCUGGCCAUGGAUGUCACCGCAAACGGUGACGGGCGUGCUCACGGGCAGGAUGUUCGUCUCCUCCAGGAGGAUG ..............................((((.((......(((((((....)))))))(((....)))...)))))).......(((((....)))))... (-23.44 = -24.25 + 0.81)
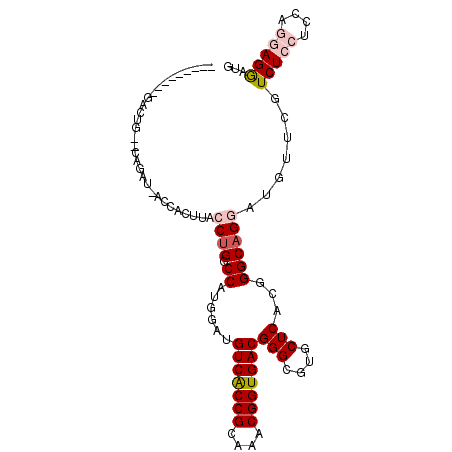

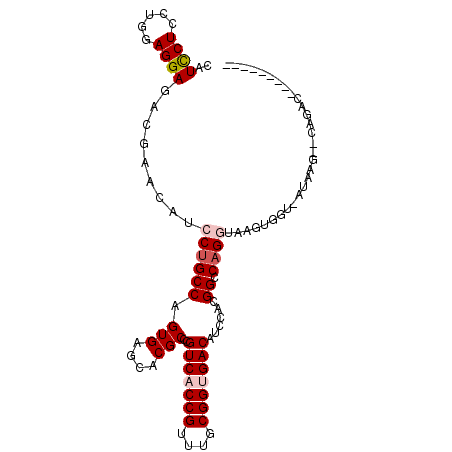
| Location | 6,217,856 – 6,217,960 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.91 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -20.56 |
| Energy contribution | -21.25 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6217856 104 - 22224390 UAUCCUCCUGGAGGAGACGAACAUCCUGCCCGUGAGCACGCCCGUCACCGUUUGCGGUGACAUCCAUGGCCAGGUAAGCGGUCAUCUCCUCCGUCUUCGUCUCC ..(((....)))(((((((((...((((.(((((.(....)..(((((((....)))))))...))))).))))...((((.........)))).))))))))) ( -43.50) >DroEre_CAF1 3005 97 - 1 CAUCCUCCUGGAGGAGACGAACAUCCUGCCCGUGAGCACGCCCGUCACCGUUUGCGGUGACAUCCAUGGCCAGGUAAGUGGUUAUUGG--CAGUCUCC-----C ..(((....)))(((((((.....).((((.((((.(((..(((((((((....)))))))..(....)...))...))).)))).))--))))))))-----. ( -38.50) >DroWil_CAF1 6630 89 - 1 CAUUCUAUUGGAGGAGACCAACAUACAGCCAGUGAGCACGCCCGUCACCGUCUGCGGUGACAUACACGGUCAGGUUAGUU------AG--CAGAACG------- ..((((.((((..((((((........((......))......(((((((....)))))))......))))...))..))------))--.))))..------- ( -22.60) >DroYak_CAF1 3145 92 - 1 CAUCCUCCUGGAGGAGACGAACAUCCUGCCCGUGAGCACGCCCGUCACCGUUUGCGGUGACAUCCAUGGUCAGGUAAGCGAUCAUCCAUCCA------------ .........((((((((((.....((((.(((((.(....)..(((((((....)))))))...))))).))))....)).)).))).))).------------ ( -34.90) >DroAna_CAF1 3801 89 - 1 CAUCCUGAUUGAGGAGACAAACAUCCUGCCAGUGAGCACGCCCGUCACCGUGUGCGGCGACAUCCACGGACAGGUGAGAUAU---AUA--CAUA---------- ..((((.....))))......((.((((((.(((.((((((........))))))(....)...))))).))))))......---...--....---------- ( -24.80) >DroPer_CAF1 4102 97 - 1 CAUCCUCCUGGAGGAGACGAACAUACUGCCAGUGAGCACGCCCGUCACCGUCUGCGGCGACAUACACGGCCAGGUGAGUGGCGCCAAG--CAGCCAGC-----C .......((((.(....).......((((...((.((..(((((((.(((....))).)))...(((......))).).))))))).)--))))))).-----. ( -29.90) >consensus CAUCCUCCUGGAGGAGACGAACAUCCUGCCAGUGAGCACGCCCGUCACCGUUUGCGGUGACAUCCACGGCCAGGUAAGUGGU_AUAAG__CAGAC_________ ..((((.....)))).........((((((.(((....)))..(((((((....)))))))......)).)))).............................. (-20.56 = -21.25 + 0.69)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:48 2006