
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,164,081 – 6,164,187 |
| Length | 106 |
| Max. P | 0.994457 |

| Location | 6,164,081 – 6,164,187 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.60 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6164081 106 + 22224390 UAUAUACAAUAUCCUCGUCUGCAUCUGCAGCGCUCUUUAGCAACGUACUCACUUCCACAGUUUGCGUGUGAAAGGAUUUCUCCUUGAGCGCAUUUAAUGCGAG--UGC .............(((((.(((....)))((((((....(((.((((...(((.....))).)))))))..(((((....))))))))))).......)))))--... ( -27.80) >DroSec_CAF1 2728 103 + 1 UGUAUAUAUUAUCG-----UGCAUCUGCAGCGCUCUUUAGCAUCGUACUCACUUCCACAGUUUGCGUGUGAAAGGAUUUCUCCUUGAGCGCAUUUAAUGCGAGUGUGC .............(-----..((..((((((((((....(((.((((...(((.....))).)))))))..(((((....)))))))))))......))))..))..) ( -27.40) >DroSim_CAF1 5074 103 + 1 UGUAUAUAUUAUCG-----UGCAUCUGCAGCGCUCUUUAGCACCGUACUCACUUCCACAGUUUGCGUGUGAAAGGAUUUCUCCUUGAGCGCAUUUAAUGCGAGUGUGC .............(-----..((..((((((((((.....(((((((...(((.....))).)))).))).(((((....)))))))))))......))))..))..) ( -29.00) >DroEre_CAF1 2681 82 + 1 UG--UAUGUCCUCA-----UGCAUCUGCAGCGCUCU-----------------UCCACAUUUUGCGUGUGAAAGGAUUUCACCUUGAGCGCUUUUAAUGCGAG--UGC ..--...(..(((.-----.((((..(.(((((((.-----------------..(((((.....))))).((((......))))))))))).)..)))))))--..) ( -25.60) >DroYak_CAF1 8453 99 + 1 UG--UAUAUCCUCU-----UGCAUCUGCAUUGCUCUUUAGCACCGCACUCACUUUCACAGUUUGCGUGUGAAAGGAUUUCACCUUGAGCGCUUUUAAUGCGAG--UGC ..--..........-----.(((..(((((((((....)))...((.(((((((((((((....).))))))))(....)....)))).))....))))))..--))) ( -26.70) >consensus UGUAUAUAUUAUCG_____UGCAUCUGCAGCGCUCUUUAGCACCGUACUCACUUCCACAGUUUGCGUGUGAAAGGAUUUCUCCUUGAGCGCAUUUAAUGCGAG__UGC ...................(((((.....((((((....................((((.......)))).((((......)))))))))).....)))))....... (-19.56 = -19.60 + 0.04)


| Location | 6,164,081 – 6,164,187 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
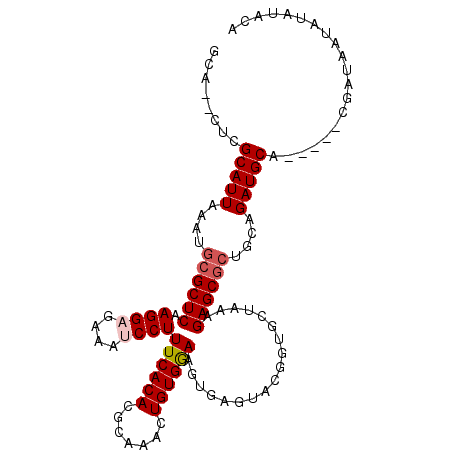
>X_DroMel_CAF1 6164081 106 - 22224390 GCA--CUCGCAUUAAAUGCGCUCAAGGAGAAAUCCUUUCACACGCAAACUGUGGAAGUGAGUACGUUGCUAAAGAGCGCUGCAGAUGCAGACGAGGAUAUUGUAUAUA ...--(((((.......(((((((((((....)))))((((.(((.....)))...)))).............))))))(((....)))).))))............. ( -31.70) >DroSec_CAF1 2728 103 - 1 GCACACUCGCAUUAAAUGCGCUCAAGGAGAAAUCCUUUCACACGCAAACUGUGGAAGUGAGUACGAUGCUAAAGAGCGCUGCAGAUGCA-----CGAUAAUAUAUACA ........(((((....(((((((((((....)))))((((.(((.....)))...)))).............))))))....))))).-----.............. ( -30.40) >DroSim_CAF1 5074 103 - 1 GCACACUCGCAUUAAAUGCGCUCAAGGAGAAAUCCUUUCACACGCAAACUGUGGAAGUGAGUACGGUGCUAAAGAGCGCUGCAGAUGCA-----CGAUAAUAUAUACA ........(((((....(((((((((((....)))))((((.(((.....)))...)))).............))))))....))))).-----.............. ( -30.40) >DroEre_CAF1 2681 82 - 1 GCA--CUCGCAUUAAAAGCGCUCAAGGUGAAAUCCUUUCACACGCAAAAUGUGGA-----------------AGAGCGCUGCAGAUGCA-----UGAGGACAUA--CA (..--((((((((...(((((((...(((((.....))))).(((.....)))..-----------------.)))))))...))))).-----.)))..)...--.. ( -28.80) >DroYak_CAF1 8453 99 - 1 GCA--CUCGCAUUAAAAGCGCUCAAGGUGAAAUCCUUUCACACGCAAACUGUGAAAGUGAGUGCGGUGCUAAAGAGCAAUGCAGAUGCA-----AGAGGAUAUA--CA (((--((.(((((....(((((((.((......))(((((((.......))))))).)))))))...(((....)))))))))).))).-----..........--.. ( -29.40) >consensus GCA__CUCGCAUUAAAUGCGCUCAAGGAGAAAUCCUUUCACACGCAAACUGUGGAAGUGAGUACGGUGCUAAAGAGCGCUGCAGAUGCA_____CGAUAAUAUAUACA ........(((((....((((((.((((....))))((((((.......))))))..................))))))....))))).................... (-23.26 = -23.90 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:28 2006