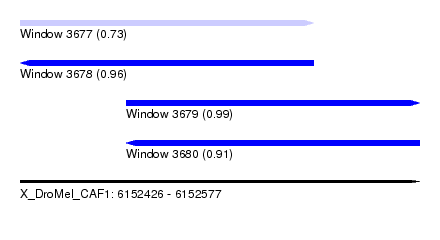
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,152,426 – 6,152,577 |
| Length | 151 |
| Max. P | 0.992305 |

| Location | 6,152,426 – 6,152,537 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.21 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -27.84 |
| Energy contribution | -27.71 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6152426 111 + 22224390 AAGUGGAAGUGGCAGUGGCAGUAGAAGUGGAGCAACAACAGCAA---------UGGCCAUGCCACAACAGUGUGGCACGGCAAGUGGAGGCCGAAGGACCCUUGAGGCAUAAUGCGCAUA ..(((...((((((.((((.......((......))..((....---------))))))))))))..((.((((.(.((((........))))(((....)))..).)))).)))))... ( -34.50) >DroSec_CAF1 5636 111 + 1 AAUUGGAAGCGGCAUUGGCAGUAGAUGUGGAGCAACAACAGCAU---------UGGCCAUGCCACAACAGUGUGGCACGGCAAGUGGAGGCCGAAGGACCCUUGAGGCAUAAUGCGCAUA ........((.((.((.(((.....))).)))).......((((---------(((((.(((((((....))))))).)))........(((.(((....)))..))).))))))))... ( -38.60) >DroEre_CAF1 3379 114 + 1 AAGUGGAAGU------GGCAGUAGAAGUGGAGCAACAGCAACAGCAACUGCGAUGGCCAUGCCACAACAGUGUGGCACGGCAAGUGGAGGCCGAAGGACCCUUGAGGCAUAAUGCGCAUA ........((------.(((((.(...((..((....))..)).).))))).)).(((.(((((((....))))))).)))..(((.(.(((.(((....)))..)))....).)))... ( -38.60) >DroYak_CAF1 2980 105 + 1 AAGUGGAAGU------GGCAGUAGAAGUGGAGCAACAACAGCAG---------UGGCCAUGCCACAACACUGUGGCACGGCAAGUGGAGGCCAAAAGACCCUUGAGGCAUAAUGCGCAUA (((.((...(------(((.......((......)).....((.---------(.(((.(((((((....))))))).))).).))...)))).....)))))...((.....))..... ( -29.00) >consensus AAGUGGAAGU______GGCAGUAGAAGUGGAGCAACAACAGCAG_________UGGCCAUGCCACAACAGUGUGGCACGGCAAGUGGAGGCCGAAGGACCCUUGAGGCAUAAUGCGCAUA ........((.......((............)).......)).............(((.(((((((....))))))).)))..(((.(.(((.(((....)))..)))....).)))... (-27.84 = -27.71 + -0.13)

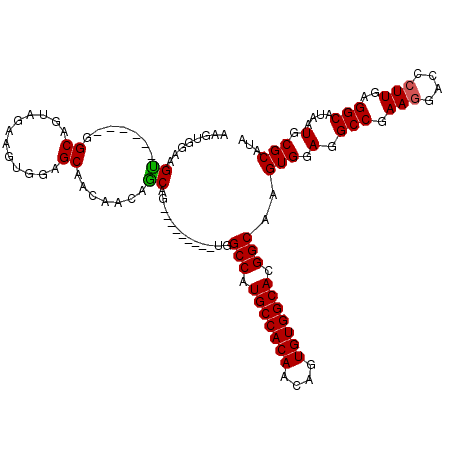
| Location | 6,152,426 – 6,152,537 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.21 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -29.29 |
| Energy contribution | -29.10 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6152426 111 - 22224390 UAUGCGCAUUAUGCCUCAAGGGUCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCA---------UUGCUGUUGUUGCUCCACUUCUACUGCCACUGCCACUUCCACUU ..((.(((....(((.....))).....(((........(((((((((((....)))))))))))..---------..((.......))............)))..)))))......... ( -30.50) >DroSec_CAF1 5636 111 - 1 UAUGCGCAUUAUGCCUCAAGGGUCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCA---------AUGCUGUUGUUGCUCCACAUCUACUGCCAAUGCCGCUUCCAAUU ...(((((((..(((.....))).....(((........(((((((((((....)))))))))))..---------..((.......))............))))))).)))........ ( -34.10) >DroEre_CAF1 3379 114 - 1 UAUGCGCAUUAUGCCUCAAGGGUCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCAUCGCAGUUGCUGUUGCUGUUGCUCCACUUCUACUGCC------ACUUCCACUU ...(((((...(((.....(((((....)))))......(((((((((((....)))))))))))....)))..)))((..((....))..))........)).------.......... ( -35.00) >DroYak_CAF1 2980 105 - 1 UAUGCGCAUUAUGCCUCAAGGGUCUUUUGGCCUCCACUUGCCGUGCCACAGUGUUGUGGCAUGGCCA---------CUGCUGUUGUUGCUCCACUUCUACUGCC------ACUUCCACUU ...(((((....((.....(((((....)))))......((((((((((......))))))))))..---------..))...)).)))...............------.......... ( -29.90) >consensus UAUGCGCAUUAUGCCUCAAGGGUCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCA_________CUGCUGUUGUUGCUCCACUUCUACUGCC______ACUUCCACUU .....(((....((..((.(((((....)))))......(((((((((((....)))))))))))............))..))...)))............................... (-29.29 = -29.10 + -0.19)

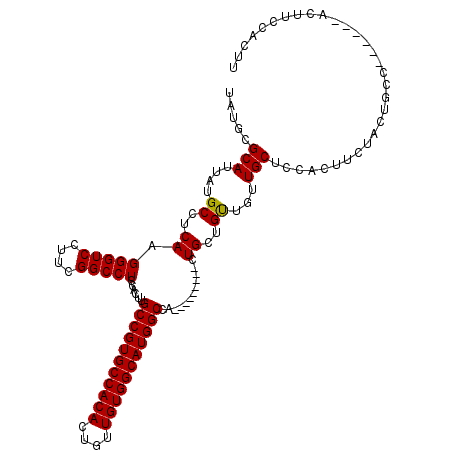
| Location | 6,152,466 – 6,152,577 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.21 |
| Mean single sequence MFE | -48.20 |
| Consensus MFE | -46.51 |
| Energy contribution | -46.33 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6152466 111 + 22224390 GCAA---------UGGCCAUGCCACAACAGUGUGGCACGGCAAGUGGAGGCCGAAGGACCCUUGAGGCAUAAUGCGCAUAUCGUUCGGUUGGCCAACCUGGCCACUGGGUCGUCCACAUC ....---------..(((.(((((((....))))))).)))..(((((((((..((((((..(((.((.......))...)))...))))((((.....)))).)).)))).)))))... ( -47.00) >DroSec_CAF1 5676 111 + 1 GCAU---------UGGCCAUGCCACAACAGUGUGGCACGGCAAGUGGAGGCCGAAGGACCCUUGAGGCAUAAUGCGCAUAUCGUUCGGUUGGCCAACCUGGCCACUGGGUCGUCCGCAUC ((((---------(((((.(((((((....))))))).)))........(((.(((....)))..))).))))))((....((((((((.((((.....)))))))))).))...))... ( -47.20) >DroEre_CAF1 3413 120 + 1 ACAGCAACUGCGAUGGCCAUGCCACAACAGUGUGGCACGGCAAGUGGAGGCCGAAGGACCCUUGAGGCAUAAUGCGCAUAUCGUUCGGUUGGCCAACCUGGCCACUGGGUCGUCCACGUC ...((....))....(((.(((((((....))))))).)))..(((((((((..((((((..(((.((.......))...)))...))))((((.....)))).)).)))).)))))... ( -48.50) >DroYak_CAF1 3014 111 + 1 GCAG---------UGGCCAUGCCACAACACUGUGGCACGGCAAGUGGAGGCCAAAAGACCCUUGAGGCAUAAUGCGCAUAUCGUUGGGUUGGCCAACCUGGCCACUGGGUCGUCCACGUC ....---------..(((.(((((((....))))))).)))..(((((((((....(((((.(((.((.......))...)))..)))))((((.....))))....)))).)))))... ( -50.10) >consensus GCAG_________UGGCCAUGCCACAACAGUGUGGCACGGCAAGUGGAGGCCGAAGGACCCUUGAGGCAUAAUGCGCAUAUCGUUCGGUUGGCCAACCUGGCCACUGGGUCGUCCACAUC ...............(((.(((((((....))))))).)))..(((((((((....((((..(((.((.......))...)))...))))((((.....))))....)))).)))))... (-46.51 = -46.33 + -0.19)



| Location | 6,152,466 – 6,152,577 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.21 |
| Mean single sequence MFE | -45.42 |
| Consensus MFE | -43.61 |
| Energy contribution | -43.68 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6152466 111 - 22224390 GAUGUGGACGACCCAGUGGCCAGGUUGGCCAACCGAACGAUAUGCGCAUUAUGCCUCAAGGGUCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCA---------UUGC ...(((((.(((((..(((((.....)))))..............((.....)).....)))))(....)..)))))..(((((((((((....)))))))))))..---------.... ( -45.80) >DroSec_CAF1 5676 111 - 1 GAUGCGGACGACCCAGUGGCCAGGUUGGCCAACCGAACGAUAUGCGCAUUAUGCCUCAAGGGUCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCA---------AUGC ((.((.((.(((((..(((((.....)))))..............((.....)).....)))))..)).)).)).....(((((((((((....)))))))))))..---------.... ( -44.40) >DroEre_CAF1 3413 120 - 1 GACGUGGACGACCCAGUGGCCAGGUUGGCCAACCGAACGAUAUGCGCAUUAUGCCUCAAGGGUCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCAUCGCAGUUGCUGU ...(((((.(((((..(((((.....)))))..............((.....)).....)))))(....)..)))))..(((((((((((....)))))))))))....((....))... ( -47.20) >DroYak_CAF1 3014 111 - 1 GACGUGGACGACCCAGUGGCCAGGUUGGCCAACCCAACGAUAUGCGCAUUAUGCCUCAAGGGUCUUUUGGCCUCCACUUGCCGUGCCACAGUGUUGUGGCAUGGCCA---------CUGC ..((....))...((((((((.((..(((((((((...((...(((.....))).))..))))....))))).)).......(((((((......))))))))))))---------))). ( -44.30) >consensus GACGUGGACGACCCAGUGGCCAGGUUGGCCAACCGAACGAUAUGCGCAUUAUGCCUCAAGGGUCCUUCGGCCUCCACUUGCCGUGCCACACUGUUGUGGCAUGGCCA_________CUGC ...(((((.(((((..(((((.....)))))..............((.....)).....)))))(....)..)))))..(((((((((((....)))))))))))............... (-43.61 = -43.68 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:18 2006