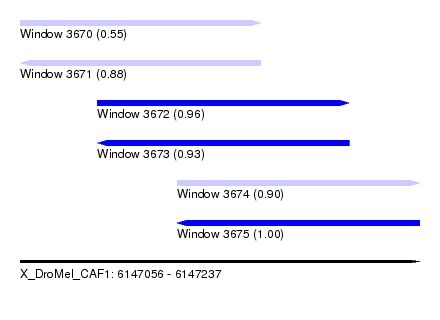
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,147,056 – 6,147,237 |
| Length | 181 |
| Max. P | 0.997194 |

| Location | 6,147,056 – 6,147,165 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 94.74 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -25.85 |
| Energy contribution | -25.63 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6147056 109 + 22224390 -----UGAUGAUGCUGCGACCGCAAUUAGCAGGCAGUGCUAAAAUUCUAACUAAUUGAUUUGCAAAAUGAUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGC -----................(((.((((((((((.((((......((((((..(((.((((((......))))))))))))))).....))))..)))....))))))).))) ( -27.60) >DroEre_CAF1 2499 114 + 1 UGCUUUGAUGAUGCUGCGACCGCAAUUAGCAGGCAGUGCUAAAACUCUACCUAAUUGAUUUGCAAAAUGAUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGU .((..(((((.(((((...((.(((((((..((.(((......)))))..))))))).((((((......)))))).......))....)))))....)))))..))....... ( -27.30) >DroYak_CAF1 2525 114 + 1 UGCUUUGAUGAUGCUGCGACCGCAAUUAGCAGGCAGUGCUAAAAUUCUACCUAAUUGAUUUGCAAAAUGAUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGCGAAAUGC .((..(((((.(((((.........((((((.....))))))........((((((..((((((......))))))...))))))....)))))....)))))..))....... ( -27.00) >consensus UGCUUUGAUGAUGCUGCGACCGCAAUUAGCAGGCAGUGCUAAAAUUCUACCUAAUUGAUUUGCAAAAUGAUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGC ............(((((((...(((((((..((.(((......)))))..)))))))..))))).(((((((((......((((.....))))...)))))))))))....... (-25.85 = -25.63 + -0.22)



| Location | 6,147,056 – 6,147,165 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 94.74 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -25.77 |
| Energy contribution | -25.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6147056 109 - 22224390 GCAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAUCAUUUUGCAAAUCAAUUAGUUAGAAUUUUAGCACUGCCUGCUAAUUGCGGUCGCAGCAUCAUCA----- (((((.......)))))...((((((..((((((((((((((((......)))))).)))..))))......((((((.....))))))....))).))))))......----- ( -29.20) >DroEre_CAF1 2499 114 - 1 ACAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAUCAUUUUGCAAAUCAAUUAGGUAGAGUUUUAGCACUGCCUGCUAAUUGCGGUCGCAGCAUCAUCAAAGCA .......(((....)))...((((((..(((....(((((((((......)))))).)))..((((((.((....)).))))))((.....))))).))))))........... ( -30.50) >DroYak_CAF1 2525 114 - 1 GCAUUUCGCAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAUCAUUUUGCAAAUCAAUUAGGUAGAAUUUUAGCACUGCCUGCUAAUUGCGGUCGCAGCAUCAUCAAAGCA ((.....(((....)))...((((((..(((....(((((((((......)))))).)))..((((((..........))))))((.....))))).))))))........)). ( -30.50) >consensus GCAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAUCAUUUUGCAAAUCAAUUAGGUAGAAUUUUAGCACUGCCUGCUAAUUGCGGUCGCAGCAUCAUCAAAGCA .......(((....)))...((((((..(((......(((((((......)))))))(((((((.(((..............)))))))))).))).))))))........... (-25.77 = -25.77 + 0.00)


| Location | 6,147,091 – 6,147,205 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 94.74 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -31.67 |
| Energy contribution | -31.57 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -4.46 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6147091 114 + 22224390 AAAAUUCUAACUAAUUGAUUUGCAAAAUGAUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGCACUUUGCCGUGAUUGGCACUCUGGCCAAUCGGAAUUUUUA ((((((((((((..(((.((((((......)))))))))))))(((((......((((((((.......)))))))))))))..(((((((......))))))))))))))).. ( -37.20) >DroEre_CAF1 2539 114 + 1 AAAACUCUACCUAAUUGAUUUGCAAAAUGAUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGUACUUUGCCGUGAUUGACACUCUUGCCGAUAGGAAUUUUUA .........(((..(((.((((((......))))))))).((((((((...(((((((((((.......))))))).)))).(((.....)))..)))))))))))........ ( -30.60) >DroYak_CAF1 2565 114 + 1 AAAAUUCUACCUAAUUGAUUUGCAAAAUGAUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGCGAAAUGCACUUUGCCGUGAUUGGCACACUGGCCAAUCGGAAUUUUUA ((((((((..((((((..((((((......))))))...))))))...((.(((((((((((.......))))))).)))).))(((((((......))))))))))))))).. ( -37.00) >consensus AAAAUUCUACCUAAUUGAUUUGCAAAAUGAUGCAAACAAAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGCACUUUGCCGUGAUUGGCACUCUGGCCAAUCGGAAUUUUUA ((((((((......(((.((((((......))))))))).((((((.....(((((((((((.......))))))).)))).(((.....)))....)))))).)))))))).. (-31.67 = -31.57 + -0.11)


| Location | 6,147,091 – 6,147,205 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 94.74 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -33.74 |
| Energy contribution | -34.63 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -5.02 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6147091 114 - 22224390 UAAAAAUUCCGAUUGGCCAGAGUGCCAAUCACGGCAAAGUGCAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAUCAUUUUGCAAAUCAAUUAGUUAGAAUUUU ..(((((((.(((((((......)))))))(((((((.(((((((.......)))))))))))))).....(((((((((((((......)))))).)))..)))).))))))) ( -38.90) >DroEre_CAF1 2539 114 - 1 UAAAAAUUCCUAUCGGCAAGAGUGUCAAUCACGGCAAAGUACAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAUCAUUUUGCAAAUCAAUUAGGUAGAGUUUU ..(((((((.((((((((((.(((.....)))(((((.((.((((.......)))).))))))).))))))....(((((((((......)))))).)))...))))))))))) ( -31.20) >DroYak_CAF1 2565 114 - 1 UAAAAAUUCCGAUUGGCCAGUGUGCCAAUCACGGCAAAGUGCAUUUCGCAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAUCAUUUUGCAAAUCAAUUAGGUAGAAUUUU ..(((((((.(((((((......)))))))(((((((.(((((((.......)))))))))))))).((((....(((((((((......)))))).)))...))))))))))) ( -41.60) >consensus UAAAAAUUCCGAUUGGCCAGAGUGCCAAUCACGGCAAAGUGCAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUUUGUUUGCAUCAUUUUGCAAAUCAAUUAGGUAGAAUUUU ..(((((((.(((((((......)))))))(((((((.(((((((.......)))))))))))))).((((....(((((((((......)))))).)))...))))))))))) (-33.74 = -34.63 + 0.89)



| Location | 6,147,127 – 6,147,237 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -22.96 |
| Energy contribution | -22.97 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6147127 110 + 22224390 CAAAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGCACUUUGCCGUGAUUGGCACUCUGGCCAAUCGGAAUUUUUAAUAUUCAUACACUAAAAACGA--------UUAGUUUUGGG .......(((((......((((((((.......))))))))))))).((((((((......))))))))((((.......)))).....((((((.(..--------...))))))). ( -29.20) >DroEre_CAF1 2575 118 + 1 CAAAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGUACUUUGCCGUGAUUGACACUCUUGCCGAUAGGAAUUUUUAAUAUUCCUCCCCUAAAUACGAGAGAGAUCGCAGUUUUGUG (((((..(((((......((((((((.......)))))))))))))((((((....((((((.....((((((.......))))))..........)))))).))))))..))))).. ( -28.96) >DroYak_CAF1 2601 105 + 1 CAAAGUUGGCAAACAGCAAGUGCAUUAUUGCGAAAUGCACUUUGCCGUGAUUGGCACACUGGCCAAUCGGAAUUUUUAAUAU-------------ACGACAGAGAUAGCAGUUUUGCC .......(((((((.(((((((((((.......))))))).)))).))(((((((......)))))))..............-------------..................))))) ( -29.90) >consensus CAAAGUUGGCAAACAGCAAGUGCAUUAUUGCUAAAUGCACUUUGCCGUGAUUGGCACUCUGGCCAAUCGGAAUUUUUAAUAUUC_U_C_CUAAA_ACGA_AGAGAU_GCAGUUUUGCG ((((..((....((.(((((((((((.......))))))).)))).))(((((((......)))))))........................................))..)))).. (-22.96 = -22.97 + 0.01)


| Location | 6,147,127 – 6,147,237 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -21.82 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6147127 110 - 22224390 CCCAAAACUAA--------UCGUUUUUAGUGUAUGAAUAUUAAAAAUUCCGAUUGGCCAGAGUGCCAAUCACGGCAAAGUGCAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUUUG .......((((--------(((((((((((((....))))))))))...))))))).((((((..(((..(((((((.(((((((.......)))))))))))))))))...)))))) ( -34.30) >DroEre_CAF1 2575 118 - 1 CACAAAACUGCGAUCUCUCUCGUAUUUAGGGGAGGAAUAUUAAAAAUUCCUAUCGGCAAGAGUGUCAAUCACGGCAAAGUACAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUUUG ........(((((......)))))..(((((.((((((.......))))))...((((((.(((.....)))(((((.((.((((.......)))).))))))).))))))..))))) ( -27.20) >DroYak_CAF1 2601 105 - 1 GGCAAAACUGCUAUCUCUGUCGU-------------AUAUUAAAAAUUCCGAUUGGCCAGUGUGCCAAUCACGGCAAAGUGCAUUUCGCAAUAAUGCACUUGCUGUUUGCCAACUUUG ((((((...((......((((((-------------..............(((((((......)))))))))))))(((((((((.......)))))))))))..))))))....... ( -35.23) >consensus CACAAAACUGC_AUCUCU_UCGU_UUUAG_G_A_GAAUAUUAAAAAUUCCGAUUGGCCAGAGUGCCAAUCACGGCAAAGUGCAUUUAGCAAUAAUGCACUUGCUGUUUGCCAACUUUG ..((((............................................(((((((......)))))))(((((((.(((((((.......))))))))))))))........)))) (-21.82 = -22.60 + 0.78)

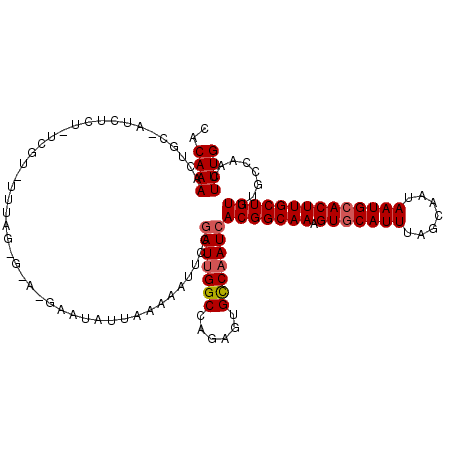

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:13 2006