
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,124,534 – 6,124,962 |
| Length | 428 |
| Max. P | 0.993279 |

| Location | 6,124,534 – 6,124,652 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.12 |
| Mean single sequence MFE | -35.11 |
| Consensus MFE | -22.38 |
| Energy contribution | -25.94 |
| Covariance contribution | 3.56 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6124534 118 + 22224390 AUAGGCA-GCUGACUGGAAAGUCGUCGCCUGCUCUCUCGCUCACUCUCGGGCGGUAGGUUACUUUGUUU-UCGCUCUCUGCCACUUAAAGUGACCGUUUCUUUUGUCUAGUCUAUUGGAA .(((((.-((.((((....)))))).))))).....((((((......))))))..((((((((((...-..((.....))....))))))))))..........(((((....))))). ( -36.30) >DroSec_CAF1 25805 119 + 1 AUAGGCAAGCUGACUGGAAAGUCGCCGCCUGCUCUCUCGCUCACUCUCGGGCGGUAGGUUACUUUGUUU-UCGCUCUCUGCCACCUAAAGUGACCGUUUCUUUUGUCUAGUCUAUUGGAA .((((((((..((((....))))((((((((................)))))))).((((((((((...-..((.....))....))))))))))......))))))))........... ( -37.89) >DroEre_CAF1 22919 119 + 1 AUAGGCAAGCUGGCUGGAAAGUCGCCGCCUGCUCUCUCGCUCACUCUCGGGCAGCUCUU-ACUUUGUUUGACAUUGUGUGCCUCUUAAUGUGGCCAUUUCUUUUGUCAAACCAAUUGAGA .(((((..((.((((....)))))).))))).......((((......))))...((((-(.((.((((((((..(((.(((.(.....).))))))......)))))))).)).))))) ( -37.70) >DroYak_CAF1 20592 120 + 1 AUAGGCAAGCUGACUGGAAAGUCGUCGCCUGCUCUCUCGCUCACUCUCGGGCAGCAGUUCACUUUGCUUGACAAUUUUUACCUCUUAAUGUGGCCAUUUCUUUUGUCUAACCUAUUGAGA .(((((..((.((((....)))))).)))))...((((((((......))))(((((......))))).(((((......((.(.....).)).........))))).........)))) ( -28.56) >consensus AUAGGCAAGCUGACUGGAAAGUCGCCGCCUGCUCUCUCGCUCACUCUCGGGCAGCAGGUUACUUUGUUU_ACACUCUCUGCCACUUAAAGUGACCAUUUCUUUUGUCUAACCUAUUGAAA .((((((((..((((....))))((((((((................)))))))).((((((((((...((.((.....)).)).))))))))))......))))))))........... (-22.38 = -25.94 + 3.56)



| Location | 6,124,652 – 6,124,771 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.46 |
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -26.20 |
| Energy contribution | -25.20 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6124652 119 + 22224390 AUGAGAUUUCGCAUUGACAAGAGUAUUUAACACAUGAAGGCCAAUCAAUUGGCUAGCUGCAAUACGCAAAAUUGGGCAUUGAAAACAAAAUGCGUAGUGCAUAUGUGCACUGCUUUUAU ..................(((((((.....((((((..((((((....))))))...((((.((((((...(((...........)))..)))))).))))))))))...))))))).. ( -29.00) >DroSec_CAF1 25924 119 + 1 AUGAGAUUUCGCAUUGGCAAGAGUAUUUAACACAUGAAGGCCAAUCAAUUGGCUAGCUGCAAUACGCAUAAUUGGGCAUUGAAAACAAAAUGCGUAGUGUAUUGGUGCACUGCUUUCAU ((((((....((((((((....((.....)).......((((((....)))))).))).((((.(.((....)).).)))).......)))))((((((((....)))))))))))))) ( -30.70) >DroEre_CAF1 23038 119 + 1 AUGAGAUUUUGCAUUGUCAAGAGUAUUUAACACAUGAAGGCCAAUCAAUUGGCUAGCUGCAAUGUGCAUAAUUAGGCAUUGAAAAUAAAAUGCGUAGUGCAUAGGUGCAAUGCUUUUAU ....(((........)))(((((((((...........((((((....))))))....(((.(((((((...((.(((((........))))).)))))))))..)))))))))))).. ( -32.20) >DroYak_CAF1 20712 119 + 1 AUGAGAUUUUGCAUUGGCAAGAGUAUUUAACAAAUGAAGACCAAUCAAUUGGUUAGCUGCAAUACGCAAAAUUAGGCAUUGAAAACAAAGUGCGUAGUGCAUUUGUGCAGUGUUUUCAU ((((((((((((....))))))................((((((....)))))).((((((....(((...(((.(((((........))))).)))))).....))))))..)))))) ( -29.70) >consensus AUGAGAUUUCGCAUUGGCAAGAGUAUUUAACACAUGAAGGCCAAUCAAUUGGCUAGCUGCAAUACGCAAAAUUAGGCAUUGAAAACAAAAUGCGUAGUGCAUAGGUGCACUGCUUUCAU ((((((....((((((.((...................((((((....)))))).(((((.....)))..((((.(((((........))))).)))))).....)))))))))))))) (-26.20 = -25.20 + -1.00)



| Location | 6,124,692 – 6,124,811 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 87.82 |
| Mean single sequence MFE | -40.27 |
| Consensus MFE | -33.26 |
| Energy contribution | -34.45 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
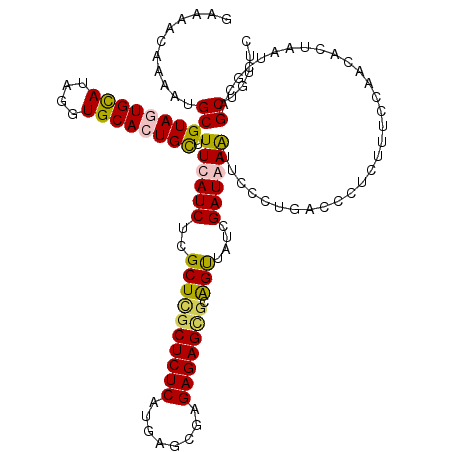
>X_DroMel_CAF1 6124692 119 + 22224390 CCAAUCAAUUGGCUAGCUGCAAUACGCAAAAUUGGGCAUUGAAAACAAAAUGCGUAGUGCAUAUGUGCACUGCUUUUAUCUCGCUCGCUCUCGUGAGCGAGAGAGCGGAGUUAUCGAUA .......(((((.(((((((((((.(((...(((.(((((........))))).)))))).))).))).((((((...((((((((((....)))))))))))))))))))))))))). ( -43.30) >DroSec_CAF1 25964 119 + 1 CCAAUCAAUUGGCUAGCUGCAAUACGCAUAAUUGGGCAUUGAAAACAAAAUGCGUAGUGUAUUGGUGCACUGCUUUCAUCUCGCUCGCUCUCAUGAGCGAGAGAGCGGAGUUAUCGAUA ...(((...(((((.((..((((((((.....((.(((((........))))).))))))))))..)).((((((...(((((((((......))))))))))))))))))))..))). ( -41.20) >DroEre_CAF1 23078 119 + 1 CCAAUCAAUUGGCUAGCUGCAAUGUGCAUAAUUAGGCAUUGAAAAUAAAAUGCGUAGUGCAUAGGUGCAAUGCUUUUAUCUCGCUCGCUCUCAUGGGCGAGAGAGUACAGUUAUCGAUA ...(((...((((((((((((.(((((((...((.(((((........))))).)))))))))..))))..))).....(((.(((((((....))))))).)))...)))))..))). ( -39.80) >DroYak_CAF1 20752 115 + 1 CCAAUCAAUUGGUUAGCUGCAAUACGCAAAAUUAGGCAUUGAAAACAAAGUGCGUAGUGCAUUUGUGCAGUGUUUUCAUCUCGCUUGCUCUCAUG----AGAGAGGGCGGCUAUCGAUU ......(((((((.((((((............(((((..((((((((..(..((.........))..)..))))))))....)))))(((((...----.))))).))))))))))))) ( -36.80) >consensus CCAAUCAAUUGGCUAGCUGCAAUACGCAAAAUUAGGCAUUGAAAACAAAAUGCGUAGUGCAUAGGUGCACUGCUUUCAUCUCGCUCGCUCUCAUGAGCGAGAGAGCGCAGUUAUCGAUA .......(((((.(((((((...............(((((........)))))((((((((....))))))))......(((.(((((((....))))))).))).)))))))))))). (-33.26 = -34.45 + 1.19)


| Location | 6,124,692 – 6,124,811 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.82 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -24.48 |
| Energy contribution | -25.73 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6124692 119 - 22224390 UAUCGAUAACUCCGCUCUCUCGCUCACGAGAGCGAGCGAGAUAAAAGCAGUGCACAUAUGCACUACGCAUUUUGUUUUCAAUGCCCAAUUUUGCGUAUUGCAGCUAGCCAAUUGAUUGG .((((((......(((.((((((((.(....).))))))))....))).(.((.....((((.((((((..(((..(.....)..)))...)))))).))))....))).))))))... ( -36.90) >DroSec_CAF1 25964 119 - 1 UAUCGAUAACUCCGCUCUCUCGCUCAUGAGAGCGAGCGAGAUGAAAGCAGUGCACCAAUACACUACGCAUUUUGUUUUCAAUGCCCAAUUAUGCGUAUUGCAGCUAGCCAAUUGAUUGG .((((((...((..(((.(((((((....))))))).)))..)).(((((((........))))((((((.(((..(.....)..)))..))))))......))).....))))))... ( -32.20) >DroEre_CAF1 23078 119 - 1 UAUCGAUAACUGUACUCUCUCGCCCAUGAGAGCGAGCGAGAUAAAAGCAUUGCACCUAUGCACUACGCAUUUUAUUUUCAAUGCCUAAUUAUGCACAUUGCAGCUAGCCAAUUGAUUGG .........((((((((.(((((.(....).))))).)))......((((((((....))))....(((((........)))))......))))....)))))....((((....)))) ( -26.50) >DroYak_CAF1 20752 115 - 1 AAUCGAUAGCCGCCCUCUCU----CAUGAGAGCAAGCGAGAUGAAAACACUGCACAAAUGCACUACGCACUUUGUUUUCAAUGCCUAAUUUUGCGUAUUGCAGCUAACCAAUUGAUUGG ((((((((((.((.(((((.----...)))))...(((((((.....((.((.((((((((.....))).)))))...)).))....))))))).....)).)))).....)))))).. ( -26.80) >consensus UAUCGAUAACUCCGCUCUCUCGCUCAUGAGAGCGAGCGAGAUAAAAGCAGUGCACAAAUGCACUACGCAUUUUGUUUUCAAUGCCCAAUUAUGCGUAUUGCAGCUAGCCAAUUGAUUGG .((((((.....(((((((........)))))))((((((((((((((((((((....))))))..)).)))))))))((((((.((....)).))))))..))).....))))))... (-24.48 = -25.73 + 1.25)


| Location | 6,124,732 – 6,124,851 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.73 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -22.62 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6124732 119 + 22224390 GAAAACAAAAUGCGUAGUGCAUAUGUGCACUGCUUUUAUCUCGCUCGCUCUCGUGAGCGAGAGAGCGGAGUUAUCGAUAAGUUGCCUGACCCUCUUUCCAACACUAAUCUCAGCUGUUG ((((.......((((((((((....)))))))).....((((((((((....))))))))))..))((.((((.(((....)))..))))))..))))(((((((......)).))))) ( -39.10) >DroSec_CAF1 26004 119 + 1 GAAAACAAAAUGCGUAGUGUAUUGGUGCACUGCUUUCAUCUCGCUCGCUCUCAUGAGCGAGAGAGCGGAGUUAUCGAUAAGUUGACGUACCCUCUUUCCAACACUGAUCUCAGCUGUGG ..........(((((....(((((((((.((((((...(((((((((......))))))))))))))).).)))))))).....)))))........(((.(((((....))).))))) ( -38.10) >DroEre_CAF1 23118 119 + 1 GAAAAUAAAAUGCGUAGUGCAUAGGUGCAAUGCUUUUAUCUCGCUCGCUCUCAUGGGCGAGAGAGUACAGUUAUCGAUAAAUUCCCUGUCCCUCUUUCCAACACUAAUCGCAGCUGUUC ....((((((.((((.(..(....)..).))))))))))(((.(((((((....))))))).))).((((((..((((............................)))).)))))).. ( -29.79) >DroYak_CAF1 20792 115 + 1 GAAAACAAAGUGCGUAGUGCAUUUGUGCAGUGUUUUCAUCUCGCUUGCUCUCAUG----AGAGAGGGCGGCUAUCGAUUAAUUCCCUGCCCCUCUUUUCAACACUAAUCGCAGCUGUUC ...((((...((((((((((((..(.((((((.........))).))).)..)))----((((.((((((.....((.....)).))))))))))......)))))..))))..)))). ( -29.90) >consensus GAAAACAAAAUGCGUAGUGCAUAGGUGCACUGCUUUCAUCUCGCUCGCUCUCAUGAGCGAGAGAGCGCAGUUAUCGAUAAAUUCCCUGACCCUCUUUCCAACACUAAUCGCAGCUGUUC ...........((((((((((....)))))))).((((((..((((((((((........))))))).)))....))))))...............................))..... (-22.62 = -22.88 + 0.25)


| Location | 6,124,851 – 6,124,962 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.70 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -16.92 |
| Energy contribution | -17.17 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6124851 111 + 22224390 -AAGUGUAUGCGUUUGUGUGUGUAUUCUAACAGAGGGAGGUGUAUAAAUUUGUAUAUACAUACAUAUGUAUGCAUGUAGACGCACAGACAUCUUCUUGUGACGUUCUUUGUU -..((((((((((.(((((((((((....(((((...(......)...)))))))))))))))).))))))))))...(((((((((........))))).))))....... ( -32.60) >DroSec_CAF1 26123 99 + 1 -AGGUGCGUGCGUGUGU-UGUGUAUUCCAACAGAGGGAGGUGUGUAAAUUUGUAUGUACAU--------AUGCAUGUAUACGCACAGACAUC---UUGUGAAGUUCUUUGUU -((((((((((((((..-((((((((((......)))).(((..((......))..))).)--------)))))...)))))))).).))))---)................ ( -25.40) >DroYak_CAF1 20907 104 + 1 C---UGCAAGUGC--GUGUGUGUAUUCUAACAGAUGGAGGUGUGUAAAUUUGUAUGUACAAAUGGAUGUACAUACAAACAAGCACGGACAUA---UUGUGACGUUCUUUGUU .---.(((((.((--((((((((.(((((.....))))).(((((...((((((((((((......))))))))))))...))))).)))))---)....))))..))))). ( -29.30) >consensus _A_GUGCAUGCGU_UGUGUGUGUAUUCUAACAGAGGGAGGUGUGUAAAUUUGUAUGUACAUA___AUGUAUGCAUGUACACGCACAGACAUC___UUGUGACGUUCUUUGUU .........(((((((((((((((((((.......))))(((((((......))))))).........)))))))))))))))....(((......)))............. (-16.92 = -17.17 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:03 2006