
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,100,532 – 6,100,633 |
| Length | 101 |
| Max. P | 0.703091 |

| Location | 6,100,532 – 6,100,633 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -16.46 |
| Energy contribution | -16.27 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
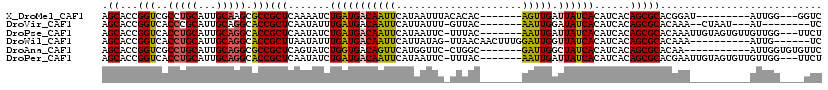
>X_DroMel_CAF1 6100532 101 + 22224390 AGCACCGGUCGCCUGCAUUGCAAGCGCCGCUCAAAAUCUGAUGACAAUUCAUAAUUUACACAC-------AGUUGAUUAUCACAUCACAGCGCACGGAU---------AUUGG---GGUC .....(((.(((.(((...))).))))))(((((.(((((((((....))))...........-------.((((............))))...)))))---------.))))---)... ( -23.20) >DroVir_CAF1 5856 99 + 1 AGCACCGGUCACCCGCAUUGCAGGCACCGCUCAAUAUUUGAUGACAAUUCAUUAUUU-GUUAC-------AAUUGGAUAUCACAUCACAGCGCACAAA--CUAAU---AU--------UC .((...(((..((.((...)).)).)))(((.((((..((((((....))))))..)-)))..-------.....(((.....)))..))))).....--.....---..--------.. ( -16.20) >DroPse_CAF1 3786 109 + 1 AGCACCGGUCACCUGCAUUGCAGGCACCGCUCAAUAUCUGAUGACAAUUCAUAAUUC-UUUAC-------AAUUGAUUAUCACAUCACAGCGCACAAAUUGUAGUGUUGUUGG---UUCU ((.((((((..(((((...))))).)))..........(((((((((((..(((...-.))).-------))))).))))))....(((((((((.....)).))))))).))---).)) ( -25.70) >DroWil_CAF1 6226 103 + 1 AGCACCGGUCACCUGCAUUGCAGGCACCGCUUAAUAUUUGAUGACAAUUCAUUAUAG-UUAACAACUUUGGAUUGGUUAUCACAUCACAGCGCACAAA----------AUUG------UC .((...(((..(((((...))))).)))(((.......(((((((((((((....((-(.....))).)))))).)))))))......))))).....----------....------.. ( -26.22) >DroAna_CAF1 3311 101 + 1 AGCACCGGUCGCCUGCAUUGCAGGCGCCGCUCAGUAUCUGGUGACAGUUCAUGGUUC-CUGGC-------GAUUGGCUAUCACAUCACAGCGCACAA-----------AUUGGUGUGUUC ....((((((((((((...))))))((((.(((....(((....)))....)))...-.))))-------))))))............(((((((..-----------....))))))). ( -36.90) >DroPer_CAF1 5526 109 + 1 AGCACCGGUCACCUGCAUUGCAGGCACCGCUCAAUAUCUGAUGACAAUUCAUAAUUC-UUUAC-------AAUUGAUUAUCACAUCACAGCGCACGAAUUGUAGUGUUGUUGG---UUCU ((.((((((..(((((...))))).)))..........(((((((((((..(((...-.))).-------))))).))))))....(((((((((.....)).))))))).))---).)) ( -25.00) >consensus AGCACCGGUCACCUGCAUUGCAGGCACCGCUCAAUAUCUGAUGACAAUUCAUAAUUC_UUUAC_______AAUUGAUUAUCACAUCACAGCGCACAAA__________AUUGG____UUC .((...(((..(((((...))))).)))(((.......(((((((((((.....................))))).))))))......)))))........................... (-16.46 = -16.27 + -0.19)

| Location | 6,100,532 – 6,100,633 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -22.30 |
| Energy contribution | -21.38 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
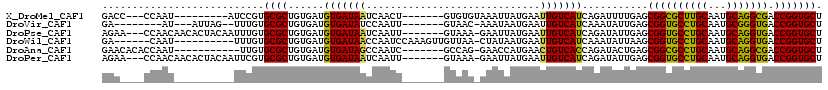
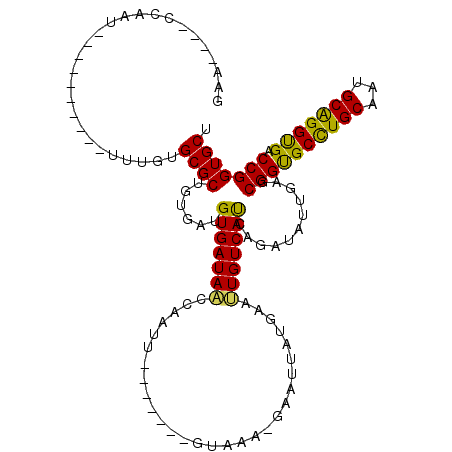
>X_DroMel_CAF1 6100532 101 - 22224390 GACC---CCAAU---------AUCCGUGCGCUGUGAUGUGAUAAUCAACU-------GUGUGUAAAUUAUGAAUUGUCAUCAGAUUUUGAGCGGCGCUUGCAAUGCAGGCGACCGGUGCU .(((---.(((.---------(((.(..(((..((((......))))...-------)))..).....((((....))))..))).)))...(((((((((...))))))).)))))... ( -27.00) >DroVir_CAF1 5856 99 - 1 GA--------AU---AUUAG--UUUGUGCGCUGUGAUGUGAUAUCCAAUU-------GUAAC-AAAUAAUGAAUUGUCAUCAAAUAUUGAGCGGUGCCUGCAAUGCGGGUGACCGGUGCU ..--------..---.....--.....(((((((...((((((....(((-------((...-..)))))....))))))...))).....((((((((((...)))))).)))))))). ( -24.70) >DroPse_CAF1 3786 109 - 1 AGAA---CCAACAACACUACAAUUUGUGCGCUGUGAUGUGAUAAUCAAUU-------GUAAA-GAAUUAUGAAUUGUCAUCAGAUAUUGAGCGGUGCCUGCAAUGCAGGUGACCGGUGCU ...(---((.....(((........))).(((...((.((((...(((((-------((((.-...)))).)))))..)))).))....)))(((((((((...)))))).))))))... ( -26.80) >DroWil_CAF1 6226 103 - 1 GA------CAAU----------UUUGUGCGCUGUGAUGUGAUAACCAAUCCAAAGUUGUUAA-CUAUAAUGAAUUGUCAUCAAAUAUUAAGCGGUGCCUGCAAUGCAGGUGACCGGUGCU ((------((((----------((.((.(((......)))...)).........(((((...-..))))))))))))).............((((((((((...)))))).))))..... ( -24.80) >DroAna_CAF1 3311 101 - 1 GAACACACCAAU-----------UUGUGCGCUGUGAUGUGAUAGCCAAUC-------GCCAG-GAACCAUGAACUGUCACCAGAUACUGAGCGGCGCCUGCAAUGCAGGCGACCGGUGCU .....((((...-----------......(((((((.(((((.....)))-------))(((-..........)))))))(((...))))))(((((((((...))))))).)))))).. ( -32.50) >DroPer_CAF1 5526 109 - 1 AGAA---CCAACAACACUACAAUUCGUGCGCUGUGAUGUGAUAAUCAAUU-------GUAAA-GAAUUAUGAAUUGUCAUCAGAUAUUGAGCGGUGCCUGCAAUGCAGGUGACCGGUGCU ....---.......(((.((((((((((..((.((((......))))...-------....)-)...))))))))))..............((((((((((...)))))).))))))).. ( -26.60) >consensus GAA____CCAAU__________UUUGUGCGCUGUGAUGUGAUAACCAAUU_______GUAAA_GAAUUAUGAAUUGUCAUCAGAUAUUGAGCGGUGCCUGCAAUGCAGGUGACCGGUGCU ...........................((((......(((((((.............................)))))))...........((((((((((...))))))).))))))). (-22.30 = -21.38 + -0.91)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:50 2006