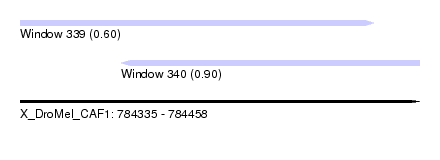
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 784,335 – 784,458 |
| Length | 123 |
| Max. P | 0.898961 |

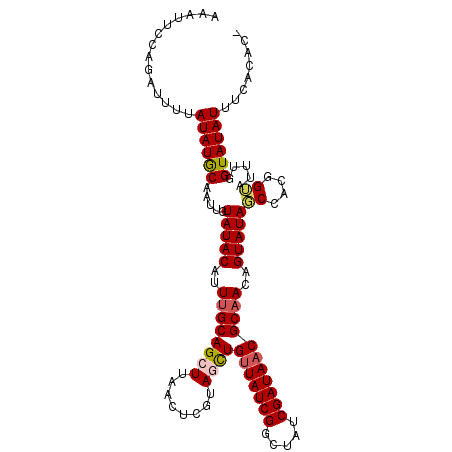
| Location | 784,335 – 784,444 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.05 |
| Mean single sequence MFE | -25.24 |
| Consensus MFE | -16.80 |
| Energy contribution | -18.16 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 784335 109 + 22224390 AAAUUCCAGAUUUUAUAUGCAAUUGUAUACAUUCGCAGCUUAACUCGUAGCUGUUAUCGGCUAUCGAUAACGCAACAGUAUAGCCAUGGUAUUUGGUAUAUUUCACACU .....((((((.((((.(((...((....))...)))(((..(((.((.((.(((((((.....))))))))).)))))..))).)))).))))))............. ( -23.60) >DroSec_CAF1 99923 107 + 1 AAAUUCCAGAUAUUAUAUGCAAUUGUAUACAUUUGCAGCUUAACUCGUACCUGCUAUCGCCUAUCGAUAACGCAACAGUAUAGCUACGGUAUUUGGUAUAUUUCACA-- .....((((((((..(((((((.((....)).)))))(((..(((.((...((((((((.....)))))..))))))))..)))))..))))))))...........-- ( -24.20) >DroSim_CAF1 61191 107 + 1 AAAUUCCAGAUAUUAUAUGCAAUUGUAUACAUUUGCAGCUUAACUCGUACCUGUUAUCGCCUAUCGAUAACGCAACAGUAUAGCUACGGUAUUUGGUAUAUUUCACA-- .....((((((((..(((((((.((....)).)))))(((..(((.((...((((((((.....))))))))..)))))..)))))..))))))))...........-- ( -27.00) >DroEre_CAF1 112712 109 + 1 AAAUUGCUGAUUUUAUAUACAGUUGUAUACAUUUGCAGCUGAACUCCUAGUUGUUAUCGGUUAUCGAUAACGCAACAGUAUAGCCUUGGGUUUUGGUAUAUUCUACACC ....(((((..........((((((((......))))))))........((.(((((((.....)))))))))..)))))..(((.........)))............ ( -25.00) >DroYak_CAF1 100585 109 + 1 AAAUCGCAGAUUUUAUAUACAGUUGUAUACAUUUGCAGCUGAACUCGUAGCUGUUAUCGGCUAUCGAUAACGCAACAGUAUAUCCUCGGGCUUUGGUAUAUUUAACACU .....((.((...(((((.((((((((......)))))))).....((.((.(((((((.....))))))))).)).)))))...))..)).................. ( -26.40) >consensus AAAUUCCAGAUUUUAUAUGCAAUUGUAUACAUUUGCAGCUUAACUCGUAGCUGUUAUCGGCUAUCGAUAACGCAACAGUAUAGCCACGGUAUUUGGUAUAUUUCACAC_ ..............((((((.....(((((..((((((((........))))(((((((.....)))))))))))..)))))((....)).....))))))........ (-16.80 = -18.16 + 1.36)

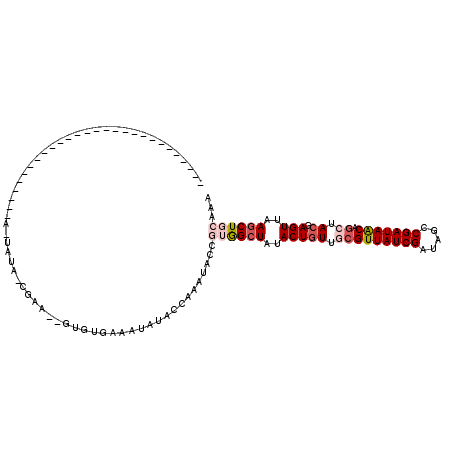
| Location | 784,366 – 784,458 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.92 |
| Mean single sequence MFE | -20.44 |
| Consensus MFE | -13.14 |
| Energy contribution | -14.54 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.898961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 784366 92 - 22224390 ------------------------GA--ACUAUAGCGAACAGUGUGAAAUAUACCAAAUACCAUGGCUAUACUGUUGCGUUAUCGAUAGCCGAUAACAGCUACGAGUUAAGCUGCGAA ------------------------.(--(((.((((.(((((((((.......(((.......))).)))))))))..(((((((.....))))))).))))..)))).......... ( -22.30) >DroSec_CAF1 99954 76 - 1 ------------------------------------------UGUGAAAUAUACCAAAUACCGUAGCUAUACUGUUGCGUUAUCGAUAGGCGAUAGCAGGUACGAGUUAAGCUGCAAA ------------------------------------------....................((((((..(((..((((((((((.....)))))))..)))..)))..))))))... ( -18.80) >DroSim_CAF1 61222 76 - 1 ------------------------------------------UGUGAAAUAUACCAAAUACCGUAGCUAUACUGUUGCGUUAUCGAUAGGCGAUAACAGGUACGAGUUAAGCUGCAAA ------------------------------------------....................((((((..(((..((((((((((.....)))))))..)))..)))..))))))... ( -18.80) >DroEre_CAF1 112743 116 - 1 AAAAUAUAUUAGAAGCAUAUAUUUGA--AAUAUACCGAAUGGUGUAGAAUAUACCAAAACCCAAGGCUAUACUGUUGCGUUAUCGAUAACCGAUAACAACUAGGAGUUCAGCUGCAAA ..............((.(((((((..--..((((((....)))))))))))))...........((((..(((.(((.(((((((.....)))))))...))).)))..))))))... ( -25.10) >DroYak_CAF1 100616 92 - 1 --------------------------GAAAUAUACCGAAUAGUGUUAAAUAUACCAAAGCCCGAGGAUAUACUGUUGCGUUAUCGAUAGCCGAUAACAGCUACGAGUUCAGCUGCAAA --------------------------...........((((((((.......................))))))))(((((((((.....)))))))((((........))))))... ( -17.20) >consensus ____________________________A_UAUA_CGAA__GUGUGAAAUAUACCAAAUACCGUGGCUAUACUGUUGCGUUAUCGAUAGCCGAUAACAGCUACGAGUUAAGCUGCAAA ..............................................................((((((..(((((.(((((((((.....))))))).)).)).)))..))))))... (-13.14 = -14.54 + 1.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:23 2006