
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,065,082 – 6,065,267 |
| Length | 185 |
| Max. P | 0.932897 |

| Location | 6,065,082 – 6,065,187 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.10 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -17.50 |
| Energy contribution | -18.75 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.702121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6065082 105 + 22224390 UCUCGAUAAUUGUUGCACGCGAUAUCGAUUGUCGCCCCAAACUUAAAGCUCUGCCCGAACUAACAGCUUUUUUCGACCAACCACCAAUAGCUGGUCACACCAGCA ..((((.((..((((...((((((.....))))))...........((.((.....)).))..))))..)).)))).............(((((.....))))). ( -24.10) >DroSec_CAF1 76007 94 + 1 UGGCGAUAAUUGUUGCAAUCGAUAUCGAUUGUCGCCCCGAACUUAAAGCUCUGC---AACUUACGGCUUUUCCCGAC-------CACUAGCUGGUCACACCGGC- .(((((((((((...(....)....)))))))))))(((.....((((((.((.---....)).))))))....(((-------((.....)))))....))).- ( -27.30) >DroSim_CAF1 72108 102 + 1 UGGCGAUAAUUGUUGCAAUCGAUAUCGAUUGUCGCCCCGAACUUAAAGCUCUGC---AACUUACGCCUUUUCCCGACCAACCACCACUAGCUGGUCACACCAGCA .((((.(((..((((((((((....))))).(((...)))............))---))))))))))......................(((((.....))))). ( -25.80) >DroYak_CAF1 76471 98 + 1 UGGCGAUUAUUGUUACCAGCGAAUUCGAUUGUCGCCAAUAACUUAAAGCUCUCC---AACUGACGGCAUUUUCCGAUU---CACCACAACCUGGUCACACU-GCG (((((((.((((.............)))).)))))))..........((.....---......(((......)))...---.((((.....))))......-)). ( -18.52) >consensus UGGCGAUAAUUGUUGCAAGCGAUAUCGAUUGUCGCCCCGAACUUAAAGCUCUGC___AACUUACGGCUUUUCCCGACC___CACCACUAGCUGGUCACACCAGCA .(((((((((((.............)))))))))))........((((((.((........)).))))))...................(((((.....))))). (-17.50 = -18.75 + 1.25)

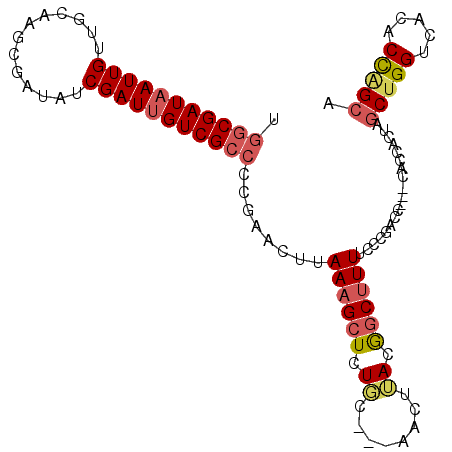
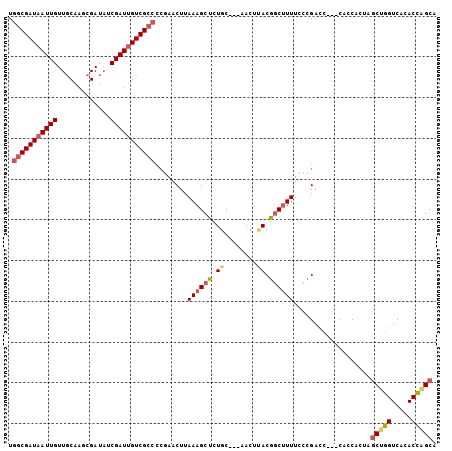
| Location | 6,065,107 – 6,065,227 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.19 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -21.04 |
| Energy contribution | -21.85 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6065107 120 + 22224390 CGAUUGUCGCCCCAAACUUAAAGCUCUGCCCGAACUAACAGCUUUUUUCGACCAACCACCAAUAGCUGGUCACACCAGCAGCAGCAAUCUUCAGCUGUUCAACAACAACAGCACGCGGCA ....((((((............(((((((..(((..((....))..)))(((((.(........).)))))......)))).)))........((((((.......))))))..)))))) ( -30.40) >DroSec_CAF1 76032 107 + 1 CGAUUGUCGCCCCGAACUUAAAGCUCUGC---AACUUACGGCUUUUCCCGAC-------CACUAGCUGGUCACACCGGC---AGCAGUCUUCAGCUGUUCAACAACAACAGCACGCGGCA ....((((((..........(((..((((---......(((......)))..-------.....(((((.....)))))---.)))).)))..((((((.......))))))..)))))) ( -32.40) >DroSim_CAF1 72133 117 + 1 CGAUUGUCGCCCCGAACUUAAAGCUCUGC---AACUUACGCCUUUUCCCGACCAACCACCACUAGCUGGUCACACCAGCAGCAGCAGUCUUCAGCUGUUCAACAACAACAGCACGCGGCA ....((((((..........(((((((((---......((........))..............(((((.....))))).)))).)).)))..((((((.......))))))..)))))) ( -31.20) >DroYak_CAF1 76496 113 + 1 CGAUUGUCGCCAAUAACUUAAAGCUCUCC---AACUGACGGCAUUUUCCGAUU---CACCACAACCUGGUCACACU-GCGCCAGCGAACUUCAGCUGUUCCACAACAACAACACGCGGCA ....((((((............((.....---......(((......)))...---.((((.....))))......-))..((((........)))).................)))))) ( -19.60) >consensus CGAUUGUCGCCCCGAACUUAAAGCUCUGC___AACUUACGGCUUUUCCCGACC___CACCACUAGCUGGUCACACCAGCAGCAGCAAUCUUCAGCUGUUCAACAACAACAGCACGCGGCA ....((((((............(((.............(((......)))..............(((((.....)))))...)))........((((((.......))))))..)))))) (-21.04 = -21.85 + 0.81)

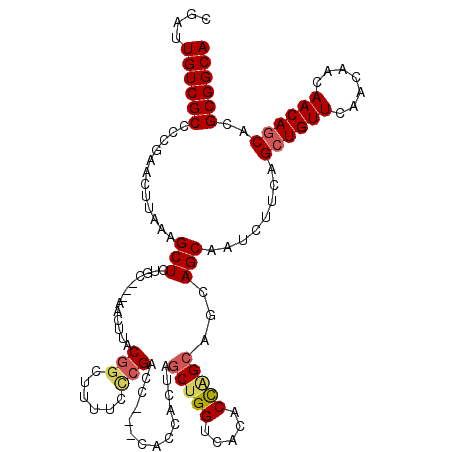
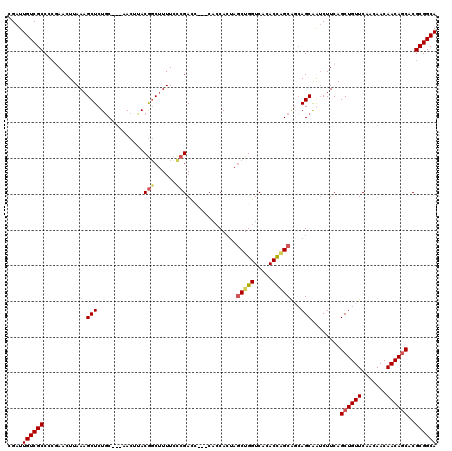
| Location | 6,065,147 – 6,065,267 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.36 |
| Mean single sequence MFE | -24.96 |
| Consensus MFE | -17.05 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6065147 120 + 22224390 GCUUUUUUCGACCAACCACCAAUAGCUGGUCACACCAGCAGCAGCAAUCUUCAGCUGUUCAACAACAACAGCACGCGGCAGCAACUACAACAAAACAUUAAGAUUGUAAUCAAGUUGUUA ............((((........(((((.....)))))....((((((((..((((((.......))))))..((....)).................))))))))......))))... ( -28.10) >DroSec_CAF1 76069 110 + 1 GCUUUUCCCGAC-------CACUAGCUGGUCACACCGGC---AGCAGUCUUCAGCUGUUCAACAACAACAGCACGCGGCAGAAACUACAACAAUACAUUAAGAUUGUAAUCAAGCUGUUA ((.......(((-------..((.(((((.....)))))---))..)))....((((((.......))))))..))(((((........(((((........))))).......))))). ( -26.76) >DroSim_CAF1 72170 120 + 1 CCUUUUCCCGACCAACCACCACUAGCUGGUCACACCAGCAGCAGCAGUCUUCAGCUGUUCAACAACAACAGCACGCGGCAGAAACUACAACAAAACAUUAAGAUUGUAAUCAAGCUGUUA ........................(((((.....)))))((((((.(((....((((((.......))))))....)))......((((((..........).))))).....)))))). ( -26.40) >DroYak_CAF1 76533 116 + 1 GCAUUUUCCGAUU---CACCACAACCUGGUCACACU-GCGCCAGCGAACUUCAGCUGUUCCACAACAACAACACGCGGCAGCGACUACAACAAAACAAUAAGAUUGUAAUCAAGUUGUUA .............---.........(((((......-..))))).((((.......))))......((((((..((....))((.((((((..........).))))).))..)))))). ( -18.60) >consensus GCUUUUCCCGACC___CACCACUAGCUGGUCACACCAGCAGCAGCAAUCUUCAGCUGUUCAACAACAACAGCACGCGGCAGAAACUACAACAAAACAUUAAGAUUGUAAUCAAGCUGUUA .........((.............(((((.....)))))....((((((((..((((((.......))))))..(..(......)..)...........))))))))..))......... (-17.05 = -17.30 + 0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:41 2006