
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,058,871 – 6,058,976 |
| Length | 105 |
| Max. P | 0.996065 |

| Location | 6,058,871 – 6,058,976 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.49 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -15.20 |
| Energy contribution | -16.53 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
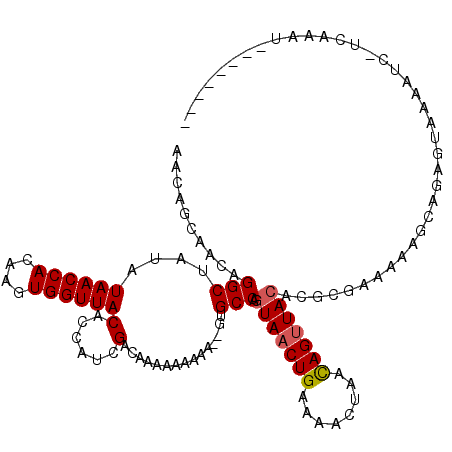
>X_DroMel_CAF1 6058871 105 + 22224390 --------AUUUGA-GAUUUUACUCCGCUUUUUCGCGUGUAACUGUUAGUUUUCAGUUACUGGCACUUUUUUUUUUUGUCGAUGGUGUAACCACUUGUGGUUAUAUAGCCUG------UU --------....((-(......)))(((......))).(((((((........))))))).(((((...........)).....(((((((((....))))))))).)))..------.. ( -24.40) >DroSec_CAF1 70069 111 + 1 --------AUUAGA-GAUUUUACUCAGCUUUUCCGCGUGUAACUGUUAGUUUUCAGUUAGUGGCACCAUUUUUUUUUGUCGGUGGUGUAACCACUUGUGGUUAUAUAGCCUGUUGCUGUU --------......-.........((((....((((...((((((........)))))))))).................(((.(((((((((....))))))))).)))....)))).. ( -28.50) >DroYak_CAF1 70201 115 + 1 CCAUACAUAUUUAAUCAAUUAAAUCUAUUUUUUUGCGUGUAUCUACUAGUUUUCAGUUACUGGCA-----UUUUUUUGUCGAUGGUGUAACCACUUGUGGUUAGAAAGCCUGUUGCUGUU ..((((((((((((....))))))............))))))...........((((.(((((((-----......)))))..(((.((((((....))))))....))).)).)))).. ( -18.90) >consensus ________AUUUGA_GAUUUUACUCAGCUUUUUCGCGUGUAACUGUUAGUUUUCAGUUACUGGCAC__UUUUUUUUUGUCGAUGGUGUAACCACUUGUGGUUAUAUAGCCUGUUGCUGUU ..................................((..(((((((........)))))))..))...................((((((((((....)))))))...))).......... (-15.20 = -16.53 + 1.33)


| Location | 6,058,871 – 6,058,976 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.49 |
| Mean single sequence MFE | -20.71 |
| Consensus MFE | -13.39 |
| Energy contribution | -13.83 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6058871 105 - 22224390 AA------CAGGCUAUAUAACCACAAGUGGUUACACCAUCGACAAAAAAAAAAAGUGCCAGUAACUGAAAACUAACAGUUACACGCGAAAAAGCGGAGUAAAAUC-UCAAAU-------- ..------..(((....((((((....))))))(......)...............))).(((((((........))))))).(((......)))(((......)-))....-------- ( -23.80) >DroSec_CAF1 70069 111 - 1 AACAGCAACAGGCUAUAUAACCACAAGUGGUUACACCACCGACAAAAAAAAAUGGUGCCACUAACUGAAAACUAACAGUUACACGCGGAAAAGCUGAGUAAAAUC-UCUAAU-------- ..((((....(((....((((((....)))))).((((..............)))))))..((((((........))))))...........)))).........-......-------- ( -23.24) >DroYak_CAF1 70201 115 - 1 AACAGCAACAGGCUUUCUAACCACAAGUGGUUACACCAUCGACAAAAAAA-----UGCCAGUAACUGAAAACUAGUAGAUACACGCAAAAAAAUAGAUUUAAUUGAUUAAAUAUGUAUGG ...(((.....)))...((((((....))))))..((((..(((......-----(((..(((.(((........))).)))..))).........((((((....)))))).))))))) ( -15.10) >consensus AACAGCAACAGGCUAUAUAACCACAAGUGGUUACACCAUCGACAAAAAAAAA__GUGCCAGUAACUGAAAACUAACAGUUACACGCGAAAAAGCAGAGUAAAAUC_UCAAAU________ ..........(((....((((((....))))))(......)...............))).(((((((........)))))))...................................... (-13.39 = -13.83 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:37 2006