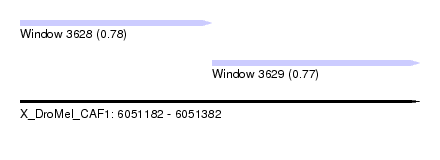
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,051,182 – 6,051,382 |
| Length | 200 |
| Max. P | 0.777471 |

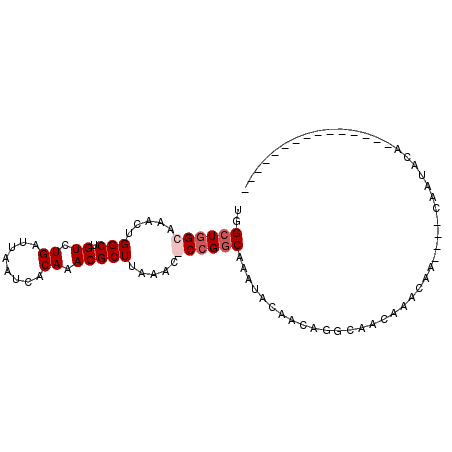
| Location | 6,051,182 – 6,051,278 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 75.63 |
| Mean single sequence MFE | -14.53 |
| Consensus MFE | -9.20 |
| Energy contribution | -10.20 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6051182 96 + 22224390 UGGCUUGCAAACUGGCUGUCUGAUUAAUCACAAACGCUUAAAU-CCGGCCAAUACAACAGGCAACAAACAACCGACAAUACACAGCACACAACACAC ..(((((.....((((((...((((...............)))-)))))))......)))))................................... ( -13.16) >DroSec_CAF1 63166 78 + 1 UGGCUGGCAAACUGGCUGUCUGAUUAAUCACAAACGCUUAAACGCCGGCAAAUACAACAGGCAACAAACAA----CAAUACA--------------- ..((((((.....(((.((.((........)).))))).....))))))..........(....)......----.......--------------- ( -16.20) >DroSim_CAF1 59084 78 + 1 UGGCUGGCAAACUGGCUGUCUGAUUAAUCACAAACGCUUAAACGCCGGCAAAUACAACAGGCAACAAACAA----CAAUACA--------------- ..((((((.....(((.((.((........)).))))).....))))))..........(....)......----.......--------------- ( -16.20) >DroYak_CAF1 61203 81 + 1 UGGCUGGCAAACUGGCUGUCUGAUUAAUCACAAACGCUUAAAU-CCGAC-------------AACAAACAACCGACAACACACU--AAGCCACACAC ((((((((......)))(((.((((...............)))-).)))-------------......................--.)))))..... ( -12.56) >consensus UGGCUGGCAAACUGGCUGUCUGAUUAAUCACAAACGCUUAAAC_CCGGCAAAUACAACAGGCAACAAACAA____CAAUACA_______________ ..((((((.....(((.((.((........)).))))).....))))))................................................ ( -9.20 = -10.20 + 1.00)


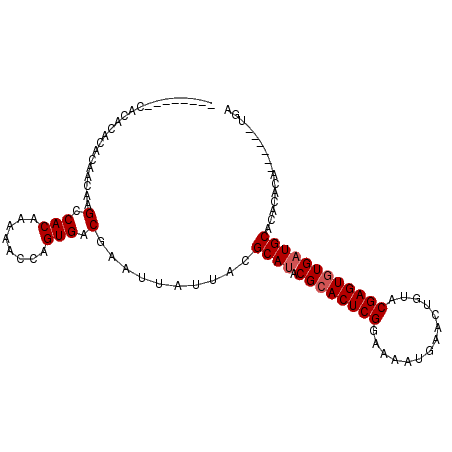
| Location | 6,051,278 – 6,051,382 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 88.04 |
| Mean single sequence MFE | -17.89 |
| Consensus MFE | -16.17 |
| Energy contribution | -16.84 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6051278 104 + 22224390 AACACACACAGACACACAACAAGCCACAAAAACCAGUGACGAAUUAUUACGCAUACGCACUCGGAAAAUGAACUGUACGAGUGUG-UGCACACACACACAUGUGA ..((((.((((...........(.(((........))).)..........((....))..............))))....(((((-((....))))))).)))). ( -20.90) >DroSec_CAF1 63244 91 + 1 --------CACACACACAACAAGCCACAAAAACGAGUGACGAAUUAUUACGCAUACGCACUCGGAAAAUGAACUGUACGAGUGUGAUGCACACACA------UGA --------..............(.(((........))).)..........((((.((((((((..............)))))))))))).......------... ( -19.54) >DroSim_CAF1 59162 91 + 1 --------CACACACACAACAAGCCACAAAAACCAGUGACGAAUUAUUACGCAUACGCACUCGGAAAAUGAACUGUACGAGUAUGAUGCACACACA------UGA --------..............(.(((........))).)..........((((.((.(((((..............))))).)))))).......------... ( -13.24) >consensus ________CACACACACAACAAGCCACAAAAACCAGUGACGAAUUAUUACGCAUACGCACUCGGAAAAUGAACUGUACGAGUGUGAUGCACACACA______UGA ......................(.(((........))).)..........((((.((((((((..............))))))))))))................ (-16.17 = -16.84 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:31 2006