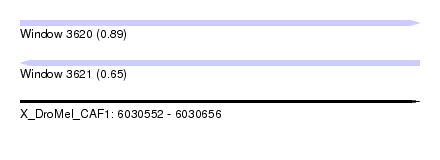
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,030,552 – 6,030,656 |
| Length | 104 |
| Max. P | 0.888180 |

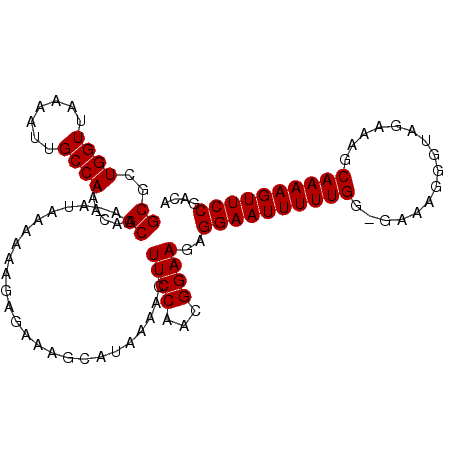
| Location | 6,030,552 – 6,030,656 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 92.97 |
| Mean single sequence MFE | -16.28 |
| Consensus MFE | -15.65 |
| Energy contribution | -16.21 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
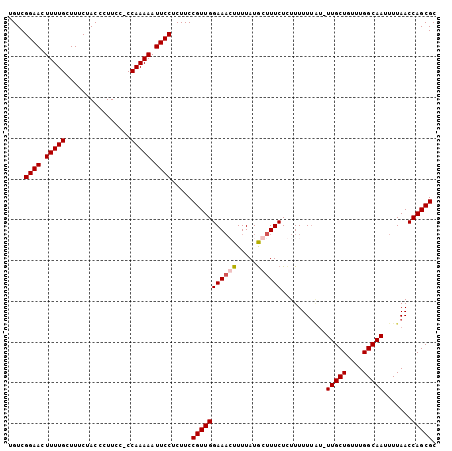
>X_DroMel_CAF1 6030552 104 + 22224390 UGUCGGAACUUUUGCUUUCUACCCUUCC-CCAAAAAUUCCUCUUCCGUUGGAAAGUUUUAUGCUUUCUCUUUUUUAUUUUGCUGUUUGGCAAUUUUAACCAGCGC ....((((.(((((..............-.))))).)))).....(((((((((((.....))))))...........(((((....))))).......))))). ( -19.36) >DroSec_CAF1 42801 104 + 1 UGUCGGAACUUUUGCUUUCUACCCUUCC-CCAAAAAUUCCACUUCCGUUGGAACGUUUUAUGCUUUCUCUUUUUUAUUUUGCUGUUUGGCAAUUUUAACCAGCGC ....((((.(((((..............-.))))).)))).....((((((...((.....))...............(((((....)))))......)))))). ( -17.16) >DroEre_CAF1 38534 101 + 1 UGUCGGAACUUUUGCUUUCUACCCUUUCCCCAAAAAUUCCUCUUCCGUUGGAAACUUUUAUACUUUCUCUUUUU----UUGCUAUUUGGCAAUUUUAACCAGCGC ....((((.(((((................))))).)))).....((((((((....))...............----(((((....)))))......)))))). ( -13.39) >DroYak_CAF1 40660 103 + 1 UGUCGGAACUUUUGCUUUCUACCCUUUU-CCAAAAAUUCCUCUUCCGUUGGAAACUUUUAUGCUUUCUUUUUUUUGU-UUGCUGUUUGGCAAUUUUAACCAGCGC ....((((.(((((..............-.))))).)))).....((((((((((....................))-))(((....)))........)))))). ( -15.21) >consensus UGUCGGAACUUUUGCUUUCUACCCUUCC_CCAAAAAUUCCUCUUCCGUUGGAAACUUUUAUGCUUUCUCUUUUUUAU_UUGCUGUUUGGCAAUUUUAACCAGCGC ....((((.(((((................))))).)))).....(((((((((((.....))))))...........(((((....))))).......))))). (-15.65 = -16.21 + 0.56)

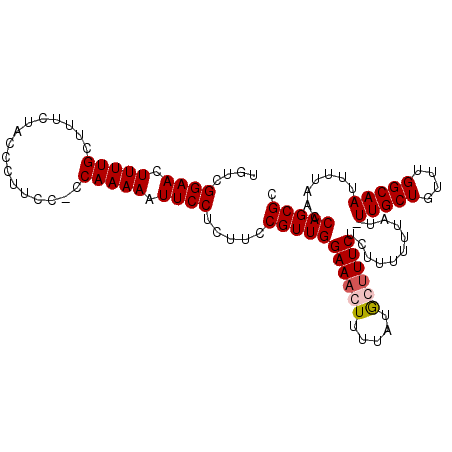
| Location | 6,030,552 – 6,030,656 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 92.97 |
| Mean single sequence MFE | -18.09 |
| Consensus MFE | -16.34 |
| Energy contribution | -16.34 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6030552 104 - 22224390 GCGCUGGUUAAAAUUGCCAAACAGCAAAAUAAAAAAGAGAAAGCAUAAAACUUUCCAACGGAAGAGGAAUUUUUGG-GGAAGGGUAGAAAGCAAAAGUUCCGACA ((..((((.......))))....))...........(.(((((.......))))))..(....).((((((((((.-..............)))))))))).... ( -19.16) >DroSec_CAF1 42801 104 - 1 GCGCUGGUUAAAAUUGCCAAACAGCAAAAUAAAAAAGAGAAAGCAUAAAACGUUCCAACGGAAGUGGAAUUUUUGG-GGAAGGGUAGAAAGCAAAAGUUCCGACA ..(((..(((...((.((.....((.................)).(((((.((((((.(....)))))))))))))-).))...)))..)))............. ( -18.93) >DroEre_CAF1 38534 101 - 1 GCGCUGGUUAAAAUUGCCAAAUAGCAA----AAAAAGAGAAAGUAUAAAAGUUUCCAACGGAAGAGGAAUUUUUGGGGAAAGGGUAGAAAGCAAAAGUUCCGACA ((..((((.......))))....))..----.....(.((((.(.....).)))))..(....).((((((((((................)))))))))).... ( -17.09) >DroYak_CAF1 40660 103 - 1 GCGCUGGUUAAAAUUGCCAAACAGCAA-ACAAAAAAAAGAAAGCAUAAAAGUUUCCAACGGAAGAGGAAUUUUUGG-AAAAGGGUAGAAAGCAAAAGUUCCGACA ((..((((.......))))....))..-..........(.((((......)))).)..(....).((((((((((.-..............)))))))))).... ( -17.16) >consensus GCGCUGGUUAAAAUUGCCAAACAGCAA_AUAAAAAAGAGAAAGCAUAAAACUUUCCAACGGAAGAGGAAUUUUUGG_GAAAGGGUAGAAAGCAAAAGUUCCGACA ((..((((.......))))....))...........................((((...))))..((((((((((................)))))))))).... (-16.34 = -16.34 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:24 2006