
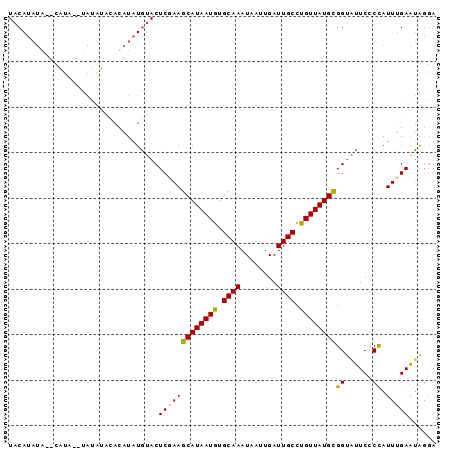
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,025,372 – 6,025,504 |
| Length | 132 |
| Max. P | 0.986140 |

| Location | 6,025,372 – 6,025,464 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 76.99 |
| Mean single sequence MFE | -19.90 |
| Consensus MFE | -12.62 |
| Energy contribution | -12.50 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6025372 92 + 22224390 UACACAUA--CACACACAUACACAUAUAUGUACUCUAAGCAUAAUGUGCAAAUAAUUGAUUGCCUGUUAUGUGGUAUUCCCCAUUUGAAUGGGA ..((((((--.(((..((..(((((.(((((.......))))))))))(((....)))..))..)))))))))......(((((....))))). ( -20.20) >DroSim_CAF1 34665 90 + 1 UACAUAUA--CAUA--UAUGUACAUAUAUGUACUCCAAGCAUAAUGUGCAAAUAAUUGAUUGCCUGUUAUGCGGUAUUCCCCAUUCGAAUGAGA ......((--((((--(((....)))))))))(((...((((((((.((((........)))).))))))))((......))........))). ( -22.40) >DroEre_CAF1 33565 76 + 1 UACAU------------------ACAUAUGUACUCGAAGCAUAAUGUGCAAAUAAUUGAUUGCCCGUUAUGCGGCACUCCCUAUUUGAGCAGUA (((((------------------....)))))((((((((((((((.((((........)))).))))))))((.....))..))))))..... ( -20.60) >DroYak_CAF1 35612 92 + 1 UACAUAUAUUCAUA--UACAUUUACAUAUAAACUCGAAGCAUAAUGUGCAAAUAAUUGAUUGCCUGUUAUGUGGUGUUCACCAUUUGAAUAGGA ......((((((..--......................((((((((.((((........)))).))))))))(((....)))...))))))... ( -16.40) >consensus UACAUAUA__CAUA__UAUAUACACAUAUGUACUCGAAGCAUAAUGUGCAAAUAAUUGAUUGCCUGUUAUGCGGUAUUCCCCAUUUGAAUAGGA .................................(((((((((((((.((((........)))).))))))))((......)).)))))...... (-12.62 = -12.50 + -0.12)


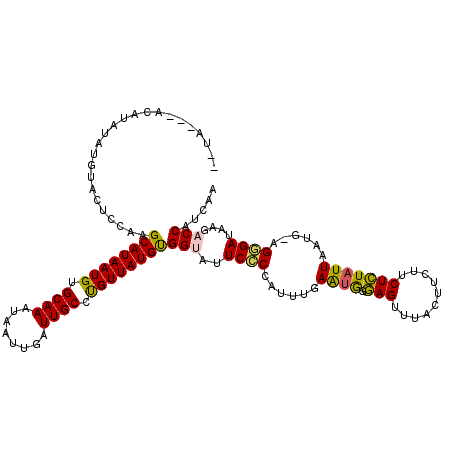
| Location | 6,025,384 – 6,025,504 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.21 |
| Mean single sequence MFE | -27.86 |
| Consensus MFE | -19.50 |
| Energy contribution | -18.94 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6025384 120 + 22224390 CACAUACACAUAUAUGUACUCUAAGCAUAAUGUGCAAAUAAUUGAUUGCCUGUUAUGUGGUAUUCCCCAUUUGAAUGGGAGUUUACUUCUUCUCUAUUAAUGCAGGGAUAAGACCAUCAA .(((((......))))).......((((((((.((((........)))).))))))))(((..((((((((.((..(((((....)))))..))....))))..))))....)))..... ( -27.60) >DroSec_CAF1 37667 113 + 1 --U----ACAUAUAUGUACUCCAAGCAUAAUGUGCAAAUAAUUGAUUGCCUGUUAUGUGGUAUUCCCCAUUCGAAUGAGAGUUUACUUCUUCUCUAUUAAUG-AGGGAUAAGACCAACAA --.----.......(((.......((((((((.((((........)))).))))))))(((..((((((((.....(((((........)))))....))))-.))))....))).))). ( -26.80) >DroSim_CAF1 34677 117 + 1 --UAUGUACAUAUAUGUACUCCAAGCAUAAUGUGCAAAUAAUUGAUUGCCUGUUAUGCGGUAUUCCCCAUUCGAAUGAGAGUUUACUUCUUCUCUAUUAAUG-AGGGAUAAGACCAUCAA --...(((((....))))).....((((((((.((((........)))).))))))))(((..((((((((.....(((((........)))))....))))-.))))....)))..... ( -32.30) >DroEre_CAF1 33570 110 + 1 ---------ACAUAUGUACUCGAAGCAUAAUGUGCAAAUAAUUGAUUGCCCGUUAUGCGGCACUCCCUAUUUGAGCAGUAGUUCACUCAUUCUGUAUUAAUG-AGCGAUAAGACCGUCAA ---------.........((((((((((((((.((((........)))).))))))))((.....))..))))))..((...((.((((((.......))))-)).))....))...... ( -25.40) >DroYak_CAF1 35626 116 + 1 --UACAUUUACAUAUAAACUCGAAGCAUAAUGUGCAAAUAAUUGAUUGCCUGUUAUGUGGUGUUCACCAUUUGAAUAGGAGUUCACUUCUUCUCUAUUAAUG-AGUGAUAUG-CCAUCAA --...................((.((((((((.((((........)))).))))))))(((((((((((((.((..(((((....)))))..))....))))-.)))).)))-)).)).. ( -27.20) >consensus __UA___ACAUAUAUGUACUCCAAGCAUAAUGUGCAAAUAAUUGAUUGCCUGUUAUGUGGUAUUCCCCAUUUGAAUGGGAGUUUACUUCUUCUCUAUUAAUG_AGGGAUAAGACCAUCAA ........................((((((((.((((........)))).))))))))(((..((((......((((.(((..........)))))))......))))....)))..... (-19.50 = -18.94 + -0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:21 2006