
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,003,722 – 6,003,830 |
| Length | 108 |
| Max. P | 0.873671 |

| Location | 6,003,722 – 6,003,830 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 71.20 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -14.74 |
| Energy contribution | -18.38 |
| Covariance contribution | 3.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6003722 108 + 22224390 GGCCGCAUUUCCCCACUUACCCACAUCCCACUCCCUUUCAGGCCAAUUGGCCGUGUCGUGGUGCACCGUAAAAUAGGCAACGCUUGUGGGGGGGAGGAGGAUGCGGGU ..................((((.(((((..((((((..((((((....)))).....(((.(((............))).)))...))..))))))..))))).)))) ( -45.80) >DroSec_CAF1 23790 103 + 1 --CCCCACUUUCCCACUUUCCCACAUCCCAUCCCCCUUCAUGCCAAUUGGCCGUGUCGCGGUGCACUGUAAAAUAGGCAACGCUUGUGGUGGGG---GGGAUGCGGGA --................((((.((((((.((((((..((((((....)).))))..(((.(((.((((...))))))).)))....)).))))---)))))).)))) ( -41.30) >DroSim_CAF1 20439 90 + 1 --CCCCACU--------UUCCCACAUCCCAUCCCCCUUCAGGCCAAUUGGCCGUGUCGCGGUGCACUGUAAAAUAGGCAACGCUUGUG-----G---GGGAUGCGGGC --.......--------........(((((((((((..((((((....)))).....(((.(((.((((...))))))).))).)).)-----)---)))))).))). ( -37.80) >DroEre_CAF1 18452 83 + 1 --CACCUU----GCACUUUGCC-------ACC--CCAACAGGCCAAUUGGCCGUGUCGCGGUGCAC-GUAAGCUAGACUCCGCUUGUGGU---------GUUGCAGGC --...(((----(((...((((-------((.--...(((((((....)))).))).((((.((..-....))......))))..)))))---------).)))))). ( -29.10) >DroYak_CAF1 20859 86 + 1 --UGCCAC----CCACAUUCCC-------ACCCUCCAUCAGGGCAAUUGGCCGUGUCGCGGUGCACUGCAAAAUAGGCAACGCUUGUGGU---------AAUGCAGGC --.(((((----((((((..((-------(((((.....))))....)))..)))).).)))(((.(((...((((((...)))))).))---------).))).))) ( -26.70) >consensus __CCCCACU___CCACUUUCCCACAUCCCACCCCCCUUCAGGCCAAUUGGCCGUGUCGCGGUGCACUGUAAAAUAGGCAACGCUUGUGGU___G____GGAUGCGGGC ...................(((.((((((...........((((....)))).....(((.(((.(((.....)))))).)))..............)))))).))). (-14.74 = -18.38 + 3.64)

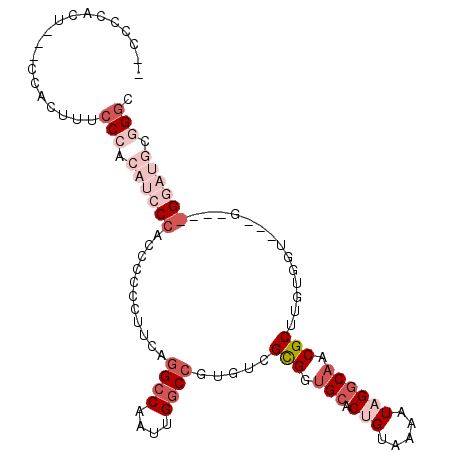
| Location | 6,003,722 – 6,003,830 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 71.20 |
| Mean single sequence MFE | -38.14 |
| Consensus MFE | -19.30 |
| Energy contribution | -22.10 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6003722 108 - 22224390 ACCCGCAUCCUCCUCCCCCCCACAAGCGUUGCCUAUUUUACGGUGCACCACGACACGGCCAAUUGGCCUGAAAGGGAGUGGGAUGUGGGUAAGUGGGGAAAUGCGGCC ((((((((((..(((((.........((((((((.......)).)))..))).((.((((....))))))...)))))..)))))))))).................. ( -42.70) >DroSec_CAF1 23790 103 - 1 UCCCGCAUCCC---CCCCACCACAAGCGUUGCCUAUUUUACAGUGCACCGCGACACGGCCAAUUGGCAUGAAGGGGGAUGGGAUGUGGGAAAGUGGGAAAGUGGGG-- (((((((((((---.(((.((....(((.(((((.......)).))).)))......(((....))).....)))))..)))))))))))................-- ( -42.60) >DroSim_CAF1 20439 90 - 1 GCCCGCAUCCC---C-----CACAAGCGUUGCCUAUUUUACAGUGCACCGCGACACGGCCAAUUGGCCUGAAGGGGGAUGGGAUGUGGGAA--------AGUGGGG-- .((((((((((---(-----(....(((.(((((.......)).))).)))..((.((((....))))))..)))))).....))))))..--------.......-- ( -38.10) >DroEre_CAF1 18452 83 - 1 GCCUGCAAC---------ACCACAAGCGGAGUCUAGCUUAC-GUGCACCGCGACACGGCCAAUUGGCCUGUUGG--GGU-------GGCAAAGUGC----AAGGUG-- (((((((.(---------((...((((........))))..-)))((((.(((((.((((....))))))))).--)))-------)......)))----.)))).-- ( -35.30) >DroYak_CAF1 20859 86 - 1 GCCUGCAUU---------ACCACAAGCGUUGCCUAUUUUGCAGUGCACCGCGACACGGCCAAUUGCCCUGAUGGAGGGU-------GGGAAUGUGG----GUGGCA-- (((..((((---------.((((..(((((((.......))))))).((.(.(((.(((.....))).)).).).))))-------)).))))..)----))....-- ( -32.00) >consensus GCCCGCAUCC____C___ACCACAAGCGUUGCCUAUUUUACAGUGCACCGCGACACGGCCAAUUGGCCUGAAGGGGGGUGGGAUGUGGGAAAGUGG___AGUGGGG__ .((((((((((..............(((.(((((.......)).))).)))..((.((((....)))))).........))))))))))................... (-19.30 = -22.10 + 2.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:18 2006