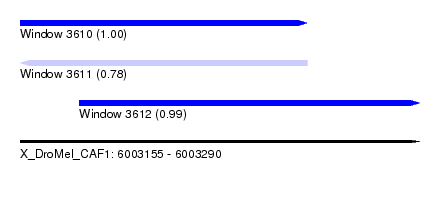
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,003,155 – 6,003,290 |
| Length | 135 |
| Max. P | 0.999971 |

| Location | 6,003,155 – 6,003,252 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -36.32 |
| Consensus MFE | -33.32 |
| Energy contribution | -33.56 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.92 |
| SVM decision value | 5.06 |
| SVM RNA-class probability | 0.999971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6003155 97 + 22224390 ---------AUUCAGGUGCUCGCUAGCAAAAACAAAGAGCACGUGCAGCACAAAGGCGGCUAAAAGUGCUUCAAAGGCAAAAGUUGGCUGUCUGUGCAGAGAGCGA ---------...((.((((((...............)))))).))..(((((..(((((((((...((((.....))))....))))))))))))))......... ( -35.06) >DroSec_CAF1 23253 101 + 1 -----GUGAACUCAGGUGCUCGCUGGCAAAAACAAAGAGCAUGUGCAGCACAAAGGCGGCUAAAAGUGCUUCAAAGGCAAAAGUUGGCUGUCUGUGCAGAGAGCGA -----((...((((.((((((..((.......))..)))))).)...(((((..(((((((((...((((.....))))....)))))))))))))).))).)).. ( -37.70) >DroSim_CAF1 19878 106 + 1 GAUGGGUGAACUCAGGUGCUCGCUGGCAAAAACAAAGAGCACGUGCAGCACAAAGGCGGCUAAAAGUGCUUCAAAGGCAAAAGUUGGCUGUCUGUGCAGAGAGCGA ..((((....)))).((((((..((.......))..))))))((.(.(((((..(((((((((...((((.....))))....)))))))))))))).)...)).. ( -40.00) >DroEre_CAF1 17953 106 + 1 GAUGGGUGGUUUCAGGUGCUCACUAGCAGAAACAAAGAGCACGAGCAGCACAAAGGCGGCGGAAAGUGCUUCAAAGGCAAAAGUUGGCUGUCUGUGCAGAGAGCGA ......((.((((..((((((...............)))))).....(((((..((((((.((...((((.....))))....)).))))))))))).)))).)). ( -35.26) >DroYak_CAF1 20280 106 + 1 GGUGGGUGGUUUCAGGUGCUCACUGGCAGAAACAAAGAGCACGUGCAGCACAAAGGAGGCUAAAAGUGCUUCAAAGGCAAAAGUUGGCUGUCUGUGCAGAGAGCGA ......((.((((..((((((..((.......))..)))))).....(((((..(..((((((...((((.....))))....))))))..)))))).)))).)). ( -33.60) >consensus G_UGGGUGAAUUCAGGUGCUCGCUGGCAAAAACAAAGAGCACGUGCAGCACAAAGGCGGCUAAAAGUGCUUCAAAGGCAAAAGUUGGCUGUCUGUGCAGAGAGCGA ..........(((..((((((..((.......))..)))))).....(((((..(((((((((...((((.....))))....)))))))))))))).)))..... (-33.32 = -33.56 + 0.24)

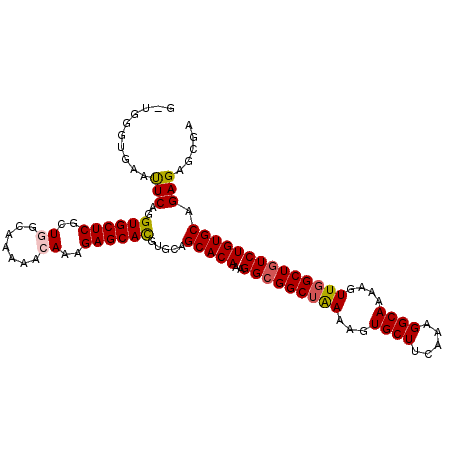
| Location | 6,003,155 – 6,003,252 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -24.39 |
| Consensus MFE | -23.46 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6003155 97 - 22224390 UCGCUCUCUGCACAGACAGCCAACUUUUGCCUUUGAAGCACUUUUAGCCGCCUUUGUGCUGCACGUGCUCUUUGUUUUUGCUAGCGAGCACCUGAAU--------- ..((((.(((((.(((((((........))......(((((...((((((....)).))))...)))))...))))).))).)).))))........--------- ( -24.80) >DroSec_CAF1 23253 101 - 1 UCGCUCUCUGCACAGACAGCCAACUUUUGCCUUUGAAGCACUUUUAGCCGCCUUUGUGCUGCACAUGCUCUUUGUUUUUGCCAGCGAGCACCUGAGUUCAC----- ..((((.(((((.((((((.................(((((..............)))))((....))...)))))).)).))).))))............----- ( -22.74) >DroSim_CAF1 19878 106 - 1 UCGCUCUCUGCACAGACAGCCAACUUUUGCCUUUGAAGCACUUUUAGCCGCCUUUGUGCUGCACGUGCUCUUUGUUUUUGCCAGCGAGCACCUGAGUUCACCCAUC ..((((.(((((.(((((((........))......(((((...((((((....)).))))...)))))...))))).)).))).))))................. ( -24.80) >DroEre_CAF1 17953 106 - 1 UCGCUCUCUGCACAGACAGCCAACUUUUGCCUUUGAAGCACUUUCCGCCGCCUUUGUGCUGCUCGUGCUCUUUGUUUCUGCUAGUGAGCACCUGAAACCACCCAUC ..((((.(((((.(((((((........))......(((((.....((.((......)).))..)))))...))))).))).)).))))................. ( -23.90) >DroYak_CAF1 20280 106 - 1 UCGCUCUCUGCACAGACAGCCAACUUUUGCCUUUGAAGCACUUUUAGCCUCCUUUGUGCUGCACGUGCUCUUUGUUUCUGCCAGUGAGCACCUGAAACCACCCACC ..((((.(((((.(((((((........))......(((((...(((((......).))))...)))))...))))).)).))).))))................. ( -25.70) >consensus UCGCUCUCUGCACAGACAGCCAACUUUUGCCUUUGAAGCACUUUUAGCCGCCUUUGUGCUGCACGUGCUCUUUGUUUUUGCCAGCGAGCACCUGAAUCCACCCA_C ..((((.(((((.(((((((........))......(((((...(((((......).))))...)))))...))))).))).)).))))................. (-23.46 = -23.70 + 0.24)

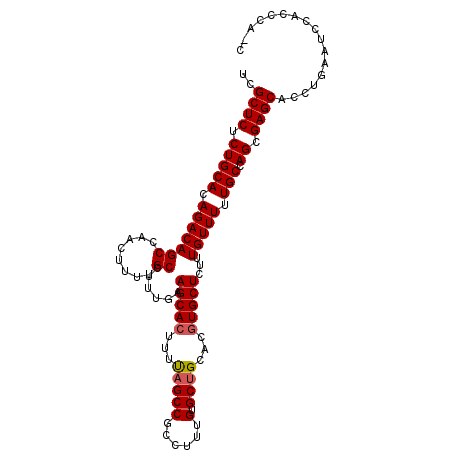

| Location | 6,003,175 – 6,003,290 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.18 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -28.92 |
| Energy contribution | -28.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.68 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6003175 115 + 22224390 AAACAAAGAGCACGUGCAGCACAAAGGCGGCUAAAAGUGCUUCAAAGGCAAAAGUUGGCUGUCUGUGCAGAGAGCGAAAAACACAAAUGCUUUAAAUACGAAAAACACACACUGC .........(((.(((..(((((..(((((((((...((((.....))))....))))))))))))))..((((((...........))))))................)))))) ( -31.20) >DroSec_CAF1 23277 100 + 1 AAACAAAGAGCAUGUGCAGCACAAAGGCGGCUAAAAGUGCUUCAAAGGCAAAAGUUGGCUGUCUGUGCAGAGAGCGAAAAACACAAAUGCUUUAAAUGCG--------------- .........((((.....(((((..(((((((((...((((.....))))....))))))))))))))..((((((...........))))))..)))).--------------- ( -31.00) >DroSim_CAF1 19907 112 + 1 AAACAAAGAGCACGUGCAGCACAAAGGCGGCUAAAAGUGCUUCAAAGGCAAAAGUUGGCUGUCUGUGCAGAGAGCGAAAAACACAAAUGCUUUAAAUGCGGGAAA---AAACUGC ......(((((((((.(.(((((..(((((((((...((((.....))))....)))))))))))))).)...)))...........))))))....((((....---...)))) ( -32.60) >consensus AAACAAAGAGCACGUGCAGCACAAAGGCGGCUAAAAGUGCUUCAAAGGCAAAAGUUGGCUGUCUGUGCAGAGAGCGAAAAACACAAAUGCUUUAAAUGCG__AAA___A_ACUGC ......(((((((((.(.(((((..(((((((((...((((.....))))....)))))))))))))).)...)))...........))))))...................... (-28.92 = -28.70 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:16 2006